Research
Technological developments in mass spectrometry
Modern mass spectrometry (MS) demands highest robustness, sensitivity, speed, and dynamic range coverage to keep up with the research community’s push towards the measurement of thousands of samples in biological and clinical settings (![]() Geyer et al., Mol. Syst. Biol., 2016,
Geyer et al., Mol. Syst. Biol., 2016, ![]() Müller et al., Nature, 2020). The Mann laboratory has a long-standing history in developing novel MS technology focusing on hardware, scan modes, and real-time MS control, with the constant goal of pushing current boundaries even further. We work in close collaboration with leading mass spectrometry companies to perform cutting-edge research on novel trapped ion mobility quadrupole time-of-flight (tims-qTOF, Bruker Daltonik GmbH), the orbitrap platform (Thermo Fisher Scientific GmbH.
Müller et al., Nature, 2020). The Mann laboratory has a long-standing history in developing novel MS technology focusing on hardware, scan modes, and real-time MS control, with the constant goal of pushing current boundaries even further. We work in close collaboration with leading mass spectrometry companies to perform cutting-edge research on novel trapped ion mobility quadrupole time-of-flight (tims-qTOF, Bruker Daltonik GmbH), the orbitrap platform (Thermo Fisher Scientific GmbH.
Milestones and recent developments
Latest milestone projects include the development of the ‘EASI-Tag’ ( Virreira Winter et al., Nat. Methods, 2018) which allows a novel way of measuring multiplexed proteomes simultaneously avoiding several pitfalls of traditional chemical labeling approaches like ratio compression. We also developed the ‘BoxCar’ method ( Meier et al., Nat. Methods, 2018), which increases the dynamic range coverage especially in challenging proteomes from body fluids or highly specialized tissues and allowed for the first time single-shot proteome measurements to a depth of more than 10.000 proteins. Currently, we are focusing on the application of a tims-qTOF to Proteomics and Metabolomics. Trapped Ion Mobility Spectrometry (TIMS) has many unique features which can be advantageous in both high-throughput and high-sensitivity settings. In TIMS, incoming ions are pushed into the device by the carrier gas facing an opposed electrical gradient with increasing field strength. As different ions have different 3D shapes in the gas-phase, they will get trapped at defined position in the TIMS device as a function of the opposed gradient, defining their Ion Mobility (IM) or collisional cross section (CCS) ( Meier et al., Nat. Comm., 2021). This allows to separate these ions into distinct species and squeeze them into very defined high-signal packages, alleviating background ion interference. Furthermore, the operating time-scale of the TIMS device fits well into the very fast sequencing speed of the qTOF exceeding a sequencing speed of more than 100 Hz and allowing for rapid scanning. On the basis of these features, we have developed the ‘Parallel Accumulation – Serial Fragmentation’ (PASEF) scan mode ( Meier et al., Mol. Cell Proteomics, 2018), characterized by extremely high fragmentation speed at undiminished sensitivity.
Intelligent ion utilization and scanning in mass spectrometry
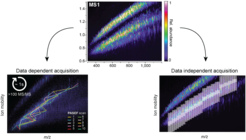
After introducing the PASEF concept on a novel TIMS-qTOF mass spectrometer, we developed a scan mode called diaPASEF ( ![]() Meier et al., Nat. Methods, 2020), which marries the data independent acquisition (DIA) strategy with PASEF, fully utilizing the properties of the TIMS device. In traditional data-independent (DIA) acquisition modes, ions are fragmented repeatedly by cycling through a pre-defined precursor m/z range with the quadrupole followed by fragmentation. This comes at a severe loss of sensitivity since generally less than 3% of the total Ion population are actually measured. In diaPASEF, which we collaborated on with the Aebersold and Roest groups, we make use of the correlation between mass-to-charge an ion mobility with up to 100% ion utilization throughout a mass spectrometry experiment. This concept is being developed further and applied to several method development projects in our laboratory. We are also currently investigating intelligent real-time data acquisition strategies, similar to the previously described ‘global targeting’ method (
Meier et al., Nat. Methods, 2020), which marries the data independent acquisition (DIA) strategy with PASEF, fully utilizing the properties of the TIMS device. In traditional data-independent (DIA) acquisition modes, ions are fragmented repeatedly by cycling through a pre-defined precursor m/z range with the quadrupole followed by fragmentation. This comes at a severe loss of sensitivity since generally less than 3% of the total Ion population are actually measured. In diaPASEF, which we collaborated on with the Aebersold and Roest groups, we make use of the correlation between mass-to-charge an ion mobility with up to 100% ion utilization throughout a mass spectrometry experiment. This concept is being developed further and applied to several method development projects in our laboratory. We are also currently investigating intelligent real-time data acquisition strategies, similar to the previously described ‘global targeting’ method ( ![]() Wichmann et al., Mol Cell Proteomics, 2019) to make scan-mode decisions based on elution profiles and precursor properties.
Wichmann et al., Mol Cell Proteomics, 2019) to make scan-mode decisions based on elution profiles and precursor properties.
Lipidomics

A comprehensive characterization of the entirety of lipids, the so-called lipidome, remains very challenging. Discriminating between lipid species that have a different chemical constitution, but the same mass-to-charge ratio is still one of the biggest bottlenecks in the field. We developed a high-sensitivity lipidomics workflow starting from minimal sample amounts based on TIMS and took advantage of PASEF to analyze precursors at more than 100 Hz while maintaining full mobility resolution of co-eluting isomers achieving attomole sensitivity (![]() Vasilopoulou et al., Nat. Commun., 2020). We are currently developing our ion mobility-enhanced four-dimensional lipidomics workflow further and are applying it to a plethora of different biological and clinical projects.
Vasilopoulou et al., Nat. Commun., 2020). We are currently developing our ion mobility-enhanced four-dimensional lipidomics workflow further and are applying it to a plethora of different biological and clinical projects.
Ultra-high sensitivity mass spectrometry and Single-Cell Proteomics
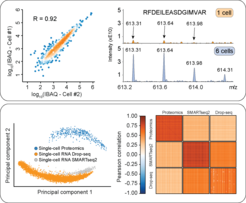
Single-cell technologies promise to revolutionize biology and precision medicine but are today mainly limited to imaging and deep sequencing technologies. However, proteins are the main drivers of cellular function and in-depth characterization of individual cells by MS-based proteomics would be highly useful and complementary. One of the major focus areas is the development of a robust single-cell proteomics workflow. It combines miniaturized sample preparation, very-low flow rate chromatography and a modified trapped ion mobility mass spectrometer (timsTOF), together resulting in hundred-fold improved sensitivity. This setup is already being applied to the analysis of single-cell and ultra-high sensitivity analyses of cell culture and tissue material with extremely low input material ( Brunner et al., MSMB, 2022).
Chromatography
Robust, reproducible, and fast chromatography is a very important building block in the MS workflow but has been somewhat neglected by the proteomics community. Since mass spectrometers have become very fast, robust, and reliable we have during the last years focused on chromatography performance ( ![]() Bache et al., Mol. Cell Proteomics, 2018). The preformed gradient concept with a relatively high flow rate of 1 µL/min already allowed us to run many large-scale projects with thousands of samples without any chromatography performance drop. Currently, we are expanding this concept, focusing on developing many versatile high-performance chromatography applications in microflow lipidomics and very low nanoflow single-cell proteomics.
Bache et al., Mol. Cell Proteomics, 2018). The preformed gradient concept with a relatively high flow rate of 1 µL/min already allowed us to run many large-scale projects with thousands of samples without any chromatography performance drop. Currently, we are expanding this concept, focusing on developing many versatile high-performance chromatography applications in microflow lipidomics and very low nanoflow single-cell proteomics.



