Research
Technological developments in mass spectrometry
Modern mass spectrometry (MS) demands highest robustness, sensitivity, speed, and dynamic range coverage to keep up with the research community’s push towards the measurement of thousands of samples in biological and clinical settings ( Geyer et al., Mol. Syst. Biol., 2016, Müller et al., Nature, 2020). The Mann laboratory has a long-standing history in developing novel MS technology focusing on hardware, scan modes, and real-time MS control, with the constant goal of pushing current boundaries even further. We work in close collaboration with leading mass spectrometry companies to perform cutting-edge research on novel trapped ion mobility quadrupole time-of-flight (timsTOF, Bruker Daltonik GmbH), the Orbitrap Astral platform (Thermo Fisher Scientific GmbH) and the ZenoTOF platform from SCIEX.
Milestones and recent developments
Milestone projects include the development of the ‘EASI-Tag’ ( Virreira Winter et al., Nat. Methods, 2018) which allows a novel way of measuring multiplexed proteomes simultaneously avoiding several pitfalls of traditional chemical labeling approaches like ratio compression. We also developed the ‘BoxCar’ method ( Meier et al., Nat. Methods, 2018), which increases the dynamic range coverage especially in challenging proteomes from body fluids or highly specialized tissues and allowed for the first time single-shot proteome measurements to a depth of more than 10.000 proteins.
Quadrupole scanning synchronized to ion mobility elution

We continuously developed novel acquisition modes for trapped ion mobility spectrometry (TIMS) and proteomics that can be advantageous in high-throughput and high-sensitivity settings.
In trapped ion mobility, incoming ions are pushed into the TIMS device by a carrier gas opposed by an electrical field gradient. As ions have different 3D shapes in the gas-phase, they become trapped at defined positions in the TIMS device, depending on their ion mobility (IM) or collisional cross section (CCS) (Meier et al., Nat. Comm., 2021).The time scale of ion mobility separation fits well with the very fast sequencing speed of the qTOF (<100 Hz). Parallel accumulation–serial fragmentation (PASEF) makes use of this speed and correlates mass-to-charge (m/z) and ion mobility separation of a TIMS-qTOF mass spectrometer (Meier et al., J. Proteome Res., 2015). With this principle, we first developed the data-dependent acquisition mode dda-PASEF, where the top N intense precursors are sampled in a ‘zig-zag’ pattern resulting in a high fragmentation speed and sensitivity (Meier et al., Mol. Cell Proteomics, 2018). In 2020, we combined data-independent acquisition with PASEF (dia-PASEF) (Meier et al., Nat. Methods, 2020), in which ions are isolated with a quadrupole selection window that cycles through a pre-defined precursor m/z range in typically two ion mobility steps. Although this could be tailored to efficiently cover the precursors with variable isolation windows (Skowronek et al., MCP, 2022), a standard dia-PASEF method did not follow the natural shape of the ion cloud. Our novel scan mode synchro-PASEF (Skowronek et al., MCP, 2023) directly links the quadrupole isolation with the ion mobility elution allowing a diagonal movement of the quadrupole and very fast cycle times. Additionally, the fast quadrupole movement with a step size below 1 Th theoretically enables the deconvolution of the highly complex data cube (Meier, Park, Mann, Mol. Cell Proteomics, 2021) to highly specific data-dependent-like ‘pure fragmentation spectra’. We recently published our accessible PASEF workflows that use the in-house developed Python tool py_diAID to automatically generate optimal dia-PASEF methods (Skowronek, Nature Protocols, 2025). Overall, every generation of a scan mode possesses increased efficiency, sensitivity and depth and is the foundation of our biological and clinical studies where we measure with minimal sample input or at high throughput.
Ultra-high sensitivity mass spectrometry and Single-Cell Proteomics
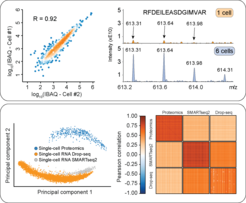
Single-cell technologies promise to revolutionize biology and precision medicine but until recently were mainly limited to imaging and deep sequencing technologies. However, proteins are the main drivers of cellular function and in-depth characterization of individual cells by MS-based proteomics would be highly useful and complementary. To that end, we recently developed a robust single-cell or ultra-high sensitive proteomics workflow. It combines miniaturized sample preparation, very-low flow rate chromatography and a very sensitive trapped ion mobility mass spectrometer (timsTOF SCP), together resulting in hundred-fold improved sensitivity (Brunner et al., MSB, 2022). Comparing the variability in single‐cell proteomes to transcriptome data revealed a stable‐core proteome despite perturbation, while the transcriptome appears stochastic.
Multiplex DIA with reference channel for enhanced sensitivity
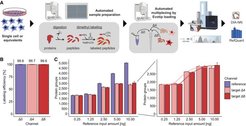
Multiplexed Data-Independent Acquisition (mDIA) is a streamlined and automated workflow that combines isotopic dimethyl labeling with DIA to enable simultaneous analysis of multiple samples with improved throughput, depth, and quantitative accuracy. (Thielert, Itang et al., Mol Syst Biol, 2023). Here, we used dimethyl labeling to multiplex samples, resulting in mass shifts of 28, 32 and 36 Da of the different channels. Since the channels are isolated from each other, one channel can be used as a reference channel with higher input material. The other two channels are used as target channels with low input material as for example single cells. The reference channel helps in identification of proteins in the target channels and increases quantitative accuracy by RefQuant, an algorithm based on the known ratio between reference channel and target channel. By using LysN, the three channels can be extended to 5-plex which doubles the throughput. We applied this to the analysis of single-cell and ultra-high sensitivity analysis of cell culture and tissue material with extremely low input (mDIA, scDVP, DVP (see below ‘clinical proteomics’)). With this, we were also able to show and prove the theory that single cells have a stable proteome.
Targeted Proteomics and Clinical Translation
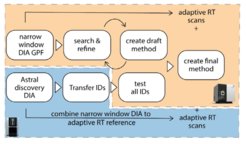
The translation of proteomics discoveries into clinical applications has long been hindered by the limitations of traditional triple quadrupole mass spectrometry technology. To address this challenge, we have developed a comprehensive strategy using the novel hybrid high-speed mass spectrometer, Stellar MS, which combines the robustness of triple quadrupoles with the enhanced capabilities of advanced linear ion trap analyzers. This platform enables extremely rapid and sensitive parallel reaction monitoring (PRM) with MS3 capabilities, allowing us to target thousands of peptides originally identified on discovery platforms like the Orbitrap Astral MS. We demonstrated seamless transfer of biomarker panels between instruments through adaptive real-time retention time correlation, achieving reproducible quantification with low coefficients of variation across the entire plasma proteome dynamic range. Furthermore, we pioneered the use of full-length ¹⁵N labeled protein standards for absolute quantification, providing enhanced accuracy and enabling detection of proteoforms and post-translational modifications. Applied to alcohol-related liver disease biomarkers, our approach showcases the potential to bridge the gap between proteomics discovery and routine clinical testing, significantly reducing assay development time from months to days (Wahle et al., bioRxiv, 2024).
Immunopeptidomics
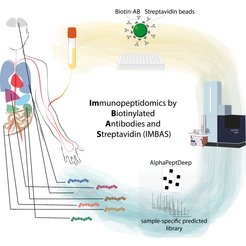
Immunopeptidomics is a cutting-edge field at the intersection of immunology and mass spectrometry, focusing on the analysis of peptides presented by major histocompatibility complex (MHC) molecules. It plays a crucial role in understanding the immune system's ability to recognize and respond to various diseases, including infections and cancers, analyzing the repertoire of peptides displayed on the cell surface. The field gained more and more interest in recent years due to its applicability and relevance in immunotherapies tailored against cancer. Our new workflow called IMBAS-MS (immunopeptidomics by biotinylated antibodies and streptavidin) combines throughput and sensitivity transferring the advances made in (single-cell) mass spectrometry towards a diverse clinical application Wahle et al., MCP, 2024)..
Chromatography
Robust, reproducible, and fast chromatography is a very important building block in the MS workflow but has been somewhat neglected by the proteomics community. Since mass spectrometers have become very fast, robust, and reliable in the last decade, we also focused on developing a chromatography platform (Bache et al., Mol. Cell Proteomics, 2018) that allowed us to run many large-scale projects with thousands of samples without any chromatography performance drop. We even expanded this concept, focusing on establishing versatile high-performance chromatography applications as nanoflow single-cell proteomics (Brunner et al., MSB, 2022) and microflow lipidomics. Advances in proteomics in the MS field can also only benefit with great chromatographic performance. We therefore collaborate with top column producers to investigate higher peak capacity and chromatographic performance on pulled emitter columns but also micro-pillar columns for low retention.





