Research
Clinical Proteomics: Body fluids and Tissues

The Clinical Proteomics teams here at the Max Planck Institute, together with our team at NNF-CPR (![]() Centre for Proteomics Research in Copenhagen) develop and apply proteomic methods for the unbiased and system-wide analysis of clinical phenotypes. We aim to systematically profile health and disease states to achieve a holistic understanding of disease mechanisms at the protein level and to uncover novel protein biomarkers for prognostic and predictive purposes. To address clinical needs and translate state-of-the-art MS methods into biomedical research, we have established a network of collaborations with multiple clinics and clinical research groups around the world, including the Medical School of the University of Chicago, Steno Diabetes Center in Denmark, Rigshospitalet Copenhagen and many more.
Centre for Proteomics Research in Copenhagen) develop and apply proteomic methods for the unbiased and system-wide analysis of clinical phenotypes. We aim to systematically profile health and disease states to achieve a holistic understanding of disease mechanisms at the protein level and to uncover novel protein biomarkers for prognostic and predictive purposes. To address clinical needs and translate state-of-the-art MS methods into biomedical research, we have established a network of collaborations with multiple clinics and clinical research groups around the world, including the Medical School of the University of Chicago, Steno Diabetes Center in Denmark, Rigshospitalet Copenhagen and many more.
Robust and reproducible methods are essential pre-requisites for clinical analyses. Using robotic assistance, we developed a blood plasma proteomics pipeline – called ‘Plasma Proteome Profiling’ – to yield a proteomic reflection of an individual’s phenotype from only 1 µl of plasma ( ![]() Geyer et al., Cell Syst., 2016). We routinely quantify more than 100 FDA-approved biomarkers and many proteins involved in important biological processes, such as proteins of the inflammation or the lipid homeostasis system. Due to its high throughput and unbiased nature, Plasma Proteome Profiling allows a paradigm shift towards a ‘rectangular’ strategy of biomarker research, in which the proteome patterns of large cohorts are correlated with their phenotypes in physiological states (
Geyer et al., Cell Syst., 2016). We routinely quantify more than 100 FDA-approved biomarkers and many proteins involved in important biological processes, such as proteins of the inflammation or the lipid homeostasis system. Due to its high throughput and unbiased nature, Plasma Proteome Profiling allows a paradigm shift towards a ‘rectangular’ strategy of biomarker research, in which the proteome patterns of large cohorts are correlated with their phenotypes in physiological states ( ![]() Geyer et al., MSB, 2017 ).
Geyer et al., MSB, 2017 ).

Recently, we implemented our expertise on liquid biopsies in the context of neurobiological disorders. Funded by the Michael J. Fox Foundation for Parkinson’s research, we have developed clinical methods for the analysis of Rab phosphorylations to measure LRRK2 activity in patient derived blood-samples ( Karayel et al., MCP, 2020). In addition, we envisioned an alternative strategy of non-invasive profiling and profiled the urinary proteome to classify mutation status and disease manifestation in patients of Parkinson’s Disease ( Virreira Winter and Karayel et al., EMBO Molecular Medicine, 2021). To describe the proteome of Alzheimer's disease (AD) in patient-derived cerebrospinal fluid (CSF), we extended our repertoire by a semi-automated workflow from CSF sample preparation to data‐independent MS acquisition. This strategy allowed us to identify consistently more than 1,000 proteins and a distinct proteomic signature for AD based on independent cohorts ( Bader et al., Mol Syst Biol., 2020).
Deep Visual Proteomics - exploring the architecture of tissues and beyond
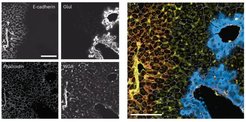
The information on the visual appearance, distribution and organization of cells is key to understanding the basic biology of nature and malfunctions such as human disease. To integrate the level of visual information into proteomic data, our group together with a multi- disciplinary team of researchers at the CPR Copenhagen and the Biological Research Centre in Szeged (Hungary, Peter Horvath Laboratory (u-szeged.hu)) developed a novel technology termed ‘Deep Visual Proteomics’ (DVP). Being the first and unique of its kind, DVP integrates advances from four different technologies for the first time into a single workflow (Mund, Coscia et al., Nature Biotechnology, 2022).

Firstly, advanced microscopy generates high-resolution tissue maps. Afterwards, machine learning and artificial intelligence (AI) algorithms are used to accurately classify cells in shape, size, or protein localization before single cells are collected by highly-accurate laser capture microdissection. After sorting normal or different types of diseased cells, the thousands of proteins present in the cell populations are detected all at once by using ultra-high sensitive mass spectrometry instruments from this minute amount of sample. Sophisticated bioinformatic analyses generate maps of proteins that provide spatial resolution of protein signatures across highly complex diseases or biological systems.
In late 2022, DVP got an extension by combining the spatial technology with our high-sensitivity single-cell proteomics workflow using multiplexed DIA (mDIA) – single-cell Deep Visual Proteomics (Rosenberger et al., Nature Methods, 2023, read also comment by Rosenberger et al., Nature Methods, 2023). We have benchmarked the promising technique on liver sections from mice, and thus characterized the heterogenous proteome of hepatocytes along the zonation axis. We currently reach a depth of 1,700 proteins per single shape, which corresponds to half or a third of a full hepatocyte. The method is ideal for characterisation of samples with very heterogeneous and sparse subtypes of cells, such as cancer stem cells.
DVP complements the efforts of our clinical proteomics team that have enabled the proteomic analysis of archived tissue biobank specimens. This includes the development of sample processing protocols for the formalin-fixed and paraffin-embedded (FFPE) tissues ( Ostasiewicz et al., J Proteome Res., 2010, Wiśniewski et al., J Proteome Res., 2011, Coscia, Doll et al., J. Pathol., 2020) In the past, this has led to the discovery of a novel prognostic biomarker in metastatic ovarian cancer ( Coscia et al., Cell, 2018) as well as identification of a master metabolic regulator fibroblast associated to high-grade serous carcinoma ( Eckert and Coscia et al., Nature, 2019). Furthermore, we have provided proof-of-concept on how a rapid and reproducible proteomic workflow can assist clinical decision-making in end-stage cancer patient ( Doll et al., Mol. Oncol., 2018, Doll, Gnad and Mann, Proteomics Clin. Appl., 2019).
Building on these results and currently applying to diverse diseases, such as melanoma, borderline ovarian cancers, rare cutaneous drug reactions, and liver diseases, together with collaboration partners in the clinics and beyond, our vision is to bring proteomics into the clinic to improve individual health by monitoring, diagnosis and treatment and – even more importantly – into daily life so that individuals stay healthy in the first place.




