Research Overview
A fundamental principle of multicellular organisms is that most cells contain the same genomic information yet represent vastly different cell types. Cellular differentiation can be artificially reversed to a pluripotent state by overexpression of the “Yamanaka factors” through a process that is inefficient and takes several days. Induced pluripotent stem cells can contribute to cell types of the embryo but not to extra-embryonic tissues. In contrast, fusion of two highly differentiated cells - egg and sperm - produces a totipotent zygote within hours and with presumably high efficiency. Totipotent cells have the potential to generate all cell types, both of the embryo and extra-embryonic tissues. The factors responsible for reprogramming to totipotency likely reside as proteins or RNA in the oocyte, since oocyte cytoplasm is sufficient to reprogram somatic cell nuclei. Despite the central importance of totipotency in establishing life, the key factors and mechanisms by which chromatin is naturally reprogrammed to a totipotent state and the triggers of zygotic genome activation (ZGA) are largely unknown in mammals.
The central aim of the department is to determine if totipotency depends on specialized reprogramming factors including pioneer transcription factors and to study their mechanisms in vivo, using mouse genetics and mechanistic cell biology, and in vitro, using biochemical assays and structural biology.
A mouse embryo followed from the zygote to the 2-cell stage by 4D time-lapse microscopy
From a transcriptionally silent zygote to the ZGA in 2-cell embryo
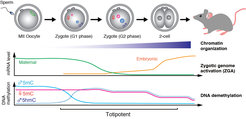
Mammalian zygotes harbour separate maternal and paternal nuclei whose genomes are contributed by the oocyte and sperm, respectively (Movie 1). After fertilisation, sperm chromatin protamines are evicted and replaced by histones to establish decondensed chromatin in the paternal nucleus. In parallel, maternal chromosomes complete meiosis II segregation, decondense and form the maternal nucleus. Zygotic maternal and paternal genomes have distinct dynamic chromatin signatures, in terms of histone and DNA modifications, which may influence their transcriptional state. Both genomes are initially silent and minor ZGA occurs mainly for paternal chromatin in G2 phase zygotes (Figure 1). Both genomes are transcriptionally activated during the major ZGA at the 2-cell stage by largely unidentified transcription factors. The embryo divides until the first lineage decision when cells of the blastocyst form either inner cell mass, which will generate the embryo, or trophectoderm, which will produce extra-embryonic tissues. Molecular details of many of these processes are poorly understood because paucity of material has made early embryos unsuitable for methods requiring large cell numbers. A revolution in low-input and single-cell approaches will now make it possible to ask mechanistic questions in zygotes.
Spatial chromatin reorganization of maternal and paternal zygotic genomes
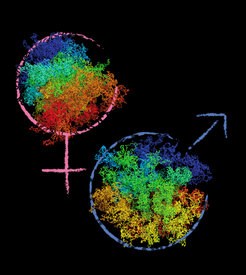
The interphase genome is organized into chromatin loops, topologically associating domains (TADs) and compartments, which reflect transcriptionally active and repressed domains. One emerging concept is that spatial chromatin organization provides a framework for regulating gene expression, which may be especially crucial for ZGA. To study zygotic genome organization, we developed chromosome conformation capture-based single-nucleus Hi-C (snHi-C) that enables the interrogation of chromatin structure in isolated zygotic maternal and paternal nuclei (Figure 2). snHi-C revealed that zygotic genomes are folded into loops and TADs but the zygotic maternal genome lacks strong compartments. By performing snHi-C on maternal cohesin knockout zygotes, we demonstrated that loops/TADs depend on the cohesin complex. This proof-of-principle experiment demonstrated that 3D chromatin structure changes of zygotes can be quantified using snHi-C. Future work will determine how chromatin organization is modulated by epigenetic reprogramming towards a totipotent chromatin “ground state”.
Epigenetic reprogramming in the early embryo
Both maternal and paternal genomes are reprogrammed on DNA and histone modifications in the early embryo. Predominantly the paternal genome undergoes active DNA demethylation. The mechanism has been elusive because methylated cytosine (5mC) can be lost via several pathways, including by passive dilution through DNA replication, or by modifications that trigger excision by an unidentified DNA glycosylase. Our molecular understanding is currently limited to a pathway involving the methylcytosine dioxygenase Tet3. A branch of Tet3-dependent DNA demethylation produces DNA breaks detected by gH2AX immunofluorescent staining of zygotes. We have provided functional genetic evidence that DNA repair is required for their timely resolution. Future work will determine how distinct mechanisms of epigenetic reprogramming contribute towards ZGA.
Mechanisms of chromatin reprogramming in vivo and in vitro
One proposed mechanism of reprogramming is by pioneer transcription factors. These are a special class of transcription factors that bind closed chromatin and either directly or indirectly displace nucleosomes, generating accessible DNA that can be bound by other proteins. Pioneer factors are essential in several cell fate transitions. The pioneer factor Zelda opens chromatin in Drosophila embryos. Future work will determine whether a set of pioneer factors for totipotency exists, analogous to the "Yamanaka factors.






