Research
A widespread mechanism regulating the functions of eukaryotic proteins involves post-translational modification by the small protein ubiquitin (UB) or structurally related ubiquitin-like proteins (UBLs).
Overview
A widespread mechanism regulating the functions of eukaryotic proteins involves post-translational modification by the small protein ubiquitin (UB) or structurally related ubiquitin-like proteins (UBLs). Covalent linkage of ubiquitin and UBLs can lead to rapid changes in a protein’s half-life, subcellular location, assembly into complexes, conformation, enzyme activities, or other properties. Moreover, defects in ubiquitin and UBL pathways are associated with numerous diseases, including cancers, neurodegenerative disorders, and viral infections. Thus, determining mechanisms underlying regulation by UB and UBLs is of broad importance for understanding numerous biological processes, and for deciphering signaling pathways and their roles in diseases.
A vast enzymatic tagging system
UB and UBL targeting is established by vast networks of hierarchical enzyme cascades, which typically involve E1 (activating), E2 (conjugating), and E3 (ligase) enzymes. The E1 first catalyzes formation of a reactive thioester-linked E2~UB or E2~UBL intermediate, which is employed by an E3 for UB or UBL linkage to specific substrates or to UB during polyubiquitylation. In humans, ubiquitylation is catalyzed by 2 E1 activating enzymes, ≈30 E2 conjugating enzymes, and more than 500 different E3 ligase enzymes, while other UBLs utilize their own distinct but related enzymes.
We are broadly interested in how UB and UBL modifications are matched with specific substrates, and how they alter the functions of their targets to regulate the cell cycle, autophagy, and other processes. We structurally visualize transient ubiquitylation complexes trapped as if in action, biochemically reconstitute novel signaling pathways, develop and use innovative chemical tools to probe ubiquitin signaling, and employ cell biology along with gene targeting to investigate how these pathways are activated precisely when needed.
Rules and regulation
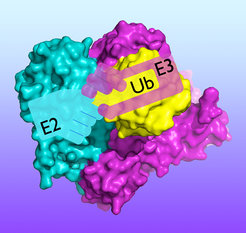
Crystal structure showing the molecular “handoff” of ubiquitin from an E2 enzyme to a HECT-type E3 enzyme.
Our early work defined structures initiating UBL conjugation, including how E1s recognize specific UBLs and promote conjugation to a cognate E2 (Walden et al. Nature 2003, Walden et al. Molecular Cell 2003, Huang et al. Nature SMB 2004, Huang et al. Molecular Cell 2005, Huang et al. Nature 2007, Huang et al. Molecular Cell 2009, Taherbhoy et al. Molecular Cell 2011, Kaiser et al. Nature SMB 2012). However, despite great importance, our knowledge of substrate modification and polyubiquitylation by E3 enzymes remains rudimentary. Thus, one focus of the department is to visualize E3 ligases trapped as if in the act of handing UB off to its recipient (Kamadurai et al. Molecular Cell 2009, Kamadurai et al. eLife 2013, Scott et al. Cell 2014, Brown et al. PNAS 2015, Brown et al. Cell 2016).
We are also studying how ubiquitylation is controlled in space and in time. We have already uncovered numerous mechanisms by which different E3 ligases are restrained until they are needed, as well as how these same E3s are activated by combinations of post-translational modifications, relief from autoinhibition, dissociation of cellular inhibitors, and/or binding to stimulatory molecules (Duda et al. Cell 2008, Scott et al. Science 2011, Duda et al. Molecular Cell 2012, Frye et al. Nature SMB 2013, Kelsall et al. EMBO J 2013, Duda et al. Structure 2013, Scott et al. Cell 2016, Yamaguchi et al. Molecular Cell 2016). Finally, we are developing novel tools to investigate the cellular and structural mechanisms by which various modifications by UB and UBLs modulate a protein’s conformations, interacting proteins, and fates (Brown et al. Molecular Cell 2014, Zhang et al. Molecular Cell 2016).
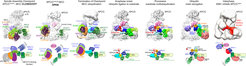
Cryo electron microscopy reconstructions of Anaphase Promoting Complex/Cyclosome, highlighting how the APC2-APC11 cullin-RING catalytic core is variously harnessed by different factors to regulate step-by-step progression through the cell cycle.
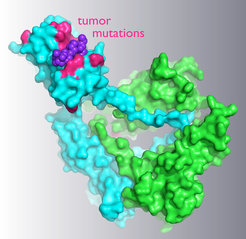
Crystal structure of a complex between the tumor suppressor E3 ligase subunit SPOP and a ubiquitylation substrate. The locations of prostate tumor-associated mutations in the substrate-binding domain are highlighted.
Ubiquitin and human health
Notably, our studies of fundamental UB conjugation mechanisms have explained how mutations in an E3 ligase disrupt ubiquitylation in many prostate cancers (Zhuang et al., Molecular Cell 2009); shown how disease-associated mutations in an E3 ligase inhibitor would lead to wayward ubiquitylation in certain vascular lesions (Duda et al., Molecular Cell 2012); revealed that some pathogenic bacteria thwart an enormous family of E3 ligases during infections (Jubelin et al., PLoS Pathogens 2010); and demonstrated that E2-E3 interactions can be selectively and effectively disrupted by small molecules (Scott et al., Nature Chemical Biology 2017).


