Research
Mechanism of the E. coli Chaperonin System, GroEL-GroES
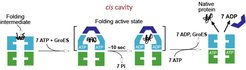
Chaperonins are large cylindrical protein complexes that assist the folding of a subset of newly-synthesized proteins in an ATP-dependent manner. They provide a compartment in which a single protein molecule is allowed to fold unimpaired by aggregation. These paradigmatic macromolecular machines are subdivided into two groups: Group I chaperonins occur in the cytosol of bacteria and within mitochondria and chloroplasts (GroEL, Hsp60s), group II chaperonins are found in the archaeal and eukaryotic cytosol (thermosome, TRiC). We have recently identified and structurally and functionally classified the substrate proteome of bacterial and archaebacterial GroEL (Kerner et al., Cell 2005). About 85 proteins were found to be strictly dependent on GroEL for folding in E. coli, including mostly proteins with complex a/b and a+b domain structures. In mechanistic work we focused on understanding how the environment of the GroEL/GroES cage affects the folding process. Specifically, we performed a mutational analysis of GroEL in which the size of the GroEL cavity and the charge properties of its wall were identified as critical features (Tang et al., Cell 2006). To better understand the mechanism of chaperonin-assisted protein folding we will employ biochemical and biophysical techniques, such as single molecule FRET and H/D exchange coupled to mass spectrometry, to define conformational transitions along the pathway of assisted protein folding.
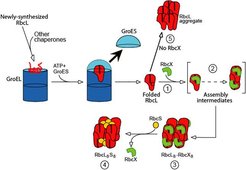
After folding, most proteins must assume a defined quarternary structure by assembling into homo- or hetero-oligomeric complexes. Little is known about chaperones that support such assembly reaction, as the primary role of the major known chaperone classes appears to be in polypeptide chain folding. To address this issue, we returned to the unresolved problem of Rubisco assembly. Rubisco, the most abundant protein on earth, is responsible for mediating almost all inorganic carbon (CO2) entry into the biosphere. Type I Rubisco, found in cyanobacteria, algae and plants, is a highly conserved hexadecameric complex consisting of 8 large subunits (~50 kDa) and 8 small subunits (~15 kDa). The simpler bacterial Rubisco (Type II) is a dimer of large subunits; it folds in a GroEL-mediated reaction and assembles spontaneously (Brinker et al., 2001). In contrast to the bacterial form, expression of Type I Rubisco (from cyanobacteria) in E. coli did not result in the production of soluble protein, whether or not GroEL/GroES was co-overexpressed. We noted that the Rubisco operon in cyanobacteria contains, besides the genes for the large and small Rubisco subunits (RbcL and RbcS, respectively), an ORF for a protein called RbcX. Very little was known about this protein, but some enhancement of Rubisco expression in E. coli upon co-expression of RbcX had been observed. Characterization of RbcX revealed that it is a homo-dimer of ~15 kDa subunits. The RbcX dimer binds to the RbcL subunits, specifically to the highly conserved C-terminal, immediately after their folding by GroEL/GroES and mediates their assembly into RbcL8 core complexes. Addition of RbcS then results in the formation of the functional holo-enzyme which does not contain RbcX. In the absence of RbcX, the RbcL subunits form non-functional aggregates (Saschenbrecker et al., Cell 2007).
Expression of Rubiscos with superior kinetic properties in plants is expected to improve the efficiency of photosynthesis, resulting in organisms with increased nitrogen and water-use efficiencies. This would improve crop performance and may also be of interest in terms of providing relief from the current increase of anthropogenically released CO2 in the atmosphere and the associated anticipated changes in global climate. Genomic data suggests that plants also contain RbcX proteins. Understanding this Rubisco-specific chaperone may thus represent a key-step towards enabling productive folding of foreign Rubiscos both in prokaryotic systems and in plants. We thus aim to address these problems by elucidating and characterizing in detail the requirements for productive Rubisco folding and assembly.
Protein Interaction and Dynamics by Mass Spectroscopy
While it has long been known that the GroEL cage can prevent the aggregation of substrate proteins during folding, it is now clear that the chaperonin actually accelerates the folding of certain substrate proteins (Tang et al., 2006). One possible explanation for this rate acceleration is that the chaperonin actively alters the preferred folding pathway of substrate proteins. This hypothesis will be tested by studying the detailed mechanism of substrate protein folding in the presence or absence of the chaperonin. A detailed study of the mechanism of folding would include the observation of the initial bound state of the substrate to the chaperonin complex, structural characterizationof specific kinetically trapped intermediate states accumulated during the folding process, and kinetic measurements of conversion between the unfolded, intermediate and folded states.
In the laboratory, the use of mass spectroscopy focuses on the identification of chaperone interaction proteomes, protein folding using hydrogen-deuterium exchange and the assembly of non-covalently bound macromolecule protein structures. The LTQ FT mass spectrometer available in the department allows the collection of very high resolution data (1 000 000 full width half maximum (FWHM) at m/z 400) at a very high mass accuracy (less than one part per million (ppm)). This instrument is therefore ideal for proteomics applications, in which high accuracy molecular weight information, combined with fragmentation spectra, allow the confident identification of peptides.
We also have a second mass spectrometer, the Waters Synapt HDMS system. This instrument contains a qTOF component, allowing us to measure a mass range up to 32 000 m/z. Since proteins and protein complexes take on large numbers of charges, this means that we can monitor complexes up to approximately one megadalton. In addition to the qTOF component, this instrument also has a Triwave cell which measures ion mobility. This additional measurement provides us with the capability of distinguishing between different conformations of the same protein complex.
With access to these two mass spectrometers with complementary capabilities, the mass spectrometry group is able to contribute to many of the projects ongoing in the Department.


