Learning from spectral experience
The final trip of many old cars leads them to a junk yard where they are deconstructed into potentially salvageable parts. By looking at the entirety of car components, a trained and experienced employee of the junk yard may be able to reconstruct the identity of the scrapped car. Mass spectrometers (MS), which identify and quantify proteins in a sample, are like molecular junk yards. The proteins are first broken into smaller fragments – the peptides. The information on the identity and the abundance of the peptides can be recorded as mass spectrometry spectra. To reconstruct the information on the proteins in the analyzed sample, the characteristics of these spectra are then compared to previously recorded libraries – a process that requires a lot of computational power.
Machine learning supports data analysis
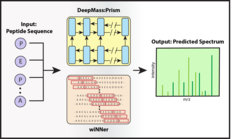
In cooperation with Verily, the life sciences company of Alphabet, researchers at the MPIB have now developed the model DeepMass:Prism to facilitate the interpretation of mass spectrometry spectra., They used machine learning to train algorithms to “translate” proteins into MS spectra. The translation of these abstract data is challenging for artificial intelligence and best performed by “deep learning” algorithms. Similar deep learning approaches are used in the automatic translation of languages. But rather than translating from English to German or vice versa, DeepMass:Prism is trained to translate between proteins and the spectra that are usually generated in MS analysis.
“The key to success in this project was the fusion of our expertise in mass spectrometry with Verily’s expertise in deep learning, particularly in the fields of biology and life sciences.”, says Jürgen Cox, independent group leader at the MPIB. Their program DeepMass:Prism was trained with more than 60 million peptide spectra from publicly accessible data bases. The program recognizes patterns from the training spectra and applies them to the analysis of new samples.
The computational biologist Cox highlights that DeepMass:Prism improves different applications of mass spectrometry. One potential use of MS is the characterization of samples whose composition is entirely unknown. The new algorithms can increase the number of peptides that are identified in this approach. Alternatively, large groups of samples with a similar general composition can be compared regarding individual differences in the protein quantity. For instance, blood samples from patients generally have a similar protein composition, but it is important to detect altered protein levels to diagnose diseases. “This is where our DeepMass:Prism has made the greatest strides”, says Cox. “Rather than experimentally determining the reference libraries to which the samples are compared, the model can now predict them – a shortcut that saves a lot of time and resources.“
Finding needles in the peptide haystack
The more than 200 cell types in the human body are characterized by the presence of different proteins, but also by the differences in the abundance of identical proteins. The different quantities of proteins are particularly challenging for MS analyses. Jürgen Cox explains the importance of measuring protein quantities with an automobile analogy: “When you completely disassemble a car, the piles of parts can look quite similar. Therefore, knowing the abundance of certain parts can help in the identification. When you find 6 cylinders in a pile, you know that it can not be a car with a four-cylinder engine.” Similarly, finding only three tires in a pile shows a potential damage of a car. The same principle can be applied to the analysis of cells or tissues. Diseases can cause certain proteins to be more or less abundant than in healthy control samples.
Many diagnostic procedures rely on the measurement of proteins in patient samples by mass spectrometry. “We need highly accurate MS to discover new biomarkers – indicators of disease. Sometimes, even a small variation of a certain biomarker can signal disease progression, therefore the prediction must be precise and reproducible.”, says Peter Cimermancic, Senior Scientist at Verily. With DeepMass:Prism, the researchers strongly improved the correlation between the predicted spectra and the actual measured spectra. He is optimistic that the model will lead to the development of new diagnostic tools.
Even though DeepMass:Prism was not trained with chemical knowledge, it discovered new chemical rules that determine how the peptides break into smaller fragments. “The previous library-based approach could only reproduce what it already knew. DeepMass:Prism is able to generate new knowledge by combining information and drawing its own conclusions. This is a very exciting finding”, says Cox, “it’s like a junk yard employee who understands where a certain part of the car is installed, even though he has never seen this type of car before. The predictions by DeepMass:Prism have led to the identification of a new kind of interaction within proteins. We believe that this discovery is only the beginning of what deep learning can do for research in life sciences.” DeepMass:Prism will be available for download on Google cloud. [CW]
Original publication
S. Tiwary*, R. Levy*, P. Gutenbrunner*, F.S. Soto, K. Palaniappan, L. Deming, M. Berndl, A. Brant, P. Cimermancic and J. Cox: High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nature Methods, May 2019 (*equal contributions)
DOI: http://dx.doi.org/10.1038/s41592-019-0427-6







