Research
The DNA origami technique allows for unprecedented control over shape and size of nanostructures. Due to their complete addressability and design controllability, DNA origami structures present a unique platform for intracellular applications such as drug delivery, to probe complex cellular signaling pathways via receptor-ligand interactions extracellularly as well as to arrange guest molecules such as nanoparticles, proteins or dyes in one, two and three dimensions.

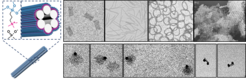
DNA origami-templated Silica nanostructures as biomedical agents
The influence of the shape of pathogens on their entry into host cells has been studied intensely. Similarly, different cellular uptake studies using synthetic nanoparticles have proven that their size and shape as well as aspect ratio and surface charge greatly influence cellular internalization pathways and efficiency in a cell-type-specific manner. To this end DNA origami presents an excellent tool for the controlled formation of discrete nanostructures with designed shape and size. However, a major bottleneck for intracellular applications of DNA origami structures is the requirement of high salt concentrations and ambient temperatures to maintain their structural integrity, as well as a susceptibility to nuclease degradation. Therefore we developed a new method to protect DNA nanostructures with a Silica coating. Silica exhibits excellent biocompatibility, non-toxicity, thermal stability, as well as chemical inertness. Using the DNA origami nanostructures as templates we can now create novel designer Silica nanostructures of various sizes and shapes. Our aim is to understand their interactions with cells (uptake, intracellular behaviour, toxicity) and proteins, esp. enzymes and to identify optimal candidates for further development into biomedical and biocatalytic agents.
Deciphering cellular signaling pathways with DNA origami
Due to their complete addressability and design controllability, DNA origami structures present a unique platform for intracellular applications such as drug delivery, but also to probe complex cellular signaling pathways via receptor-ligand interactions extracellularly. One of our research interests is to use the inherent addressability of DNA origami structures for the controlled arrangement of ligands to study interactions and signaling pathway initiations of immune cells.
One of the systems we are interested in is the initiation of apoptosis signaling by Fas ligand (FasL) - Fas receptor (FasR) interactions. It is believed that FasL, expressed on cytotocix T-cells, must arrange FasR into a hexagonal arrangement for effective apoptosis induction. By mimicking this hexagonal arrangement, as well as other geometries, of FasL on a DNA origami platform we aim to shed light on the mechanism and requirements for Fas-induced apoptosis signaling.

Enzyme cascades in silicified DNA origami structures
A common dream in biocatalysis research is the achievement of a biochemical nanofactory, where multiple components work together in order to produce new unique materials with improved functions, that can rival the efficiency of biological systems. However, for this to occur several factors need to be considered. Living organisms have developed two complementary strategies for successful multi-enzyme reactions. These are compartmentalization and substrate channeling, where substrates or intermediary products are transferred locally from one reactant to another. Similarly in artificial (multi-) enzyme reactions the enzymes must be protected from external negative factors such as proteases, pH and temperature changes. Our aim is to use silicified DNA origami structures as solid supports for the controlled arrangement of enzymes and as a controlled environment, optimized for each enzyme in order to create highly efficient enzyme cascades.
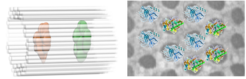
DNA origami frameworks for the structural analysis of proteins

More than three decades ago, Nadrian Seeman proposed the co-crystallization of biological macromolecules on self-assembled DNA lattices to solve their structure by X-ray crystallography. This concept has catalyzed the success of the rapidly developing area of DNA nanotechnology and recently single-crystal DNA origami crystals have been developed. Thus far, however, arranging biological macromolecules in three dimensions using DNA frameworks has not been achieved, mainly because the reported DNA lattices lack sufficient rigidity and / or cavity size to host sizeable guest molecules. To this end we developed DNA origami-based crystal lattices that offers space for up to 30 nm large particles. Our aim is to design (single-) crystalline) DNA origami host frameworks for X-ray and cryoEM investigations of protein guest molecules positioned inside the DNA origami host matrix.






