Graph Kernels
Mahito Sugiyama, Elisabetta Ghisu, Felipe Llinares-Lopez, Karsten Borgwardt

graphkernels: R and Python packages for graph comparison
Summary
R and Python packages for computing graph kernels, which include:
- simple kernels between vertex and/or edge label histograms
- random walk kernels (popular baselines)
- Weisfeiler-Lehman graph kernel (state-of-the-art)
- Shortest-Path kernel
- graphlet kernel
Code
R
Python
Reference
graphkernels: R and Python packages for graph comparison
Mahito Sugiyama, Elisabetta Ghisu, Felipe Llinares-Lopez, Karsten Borgwardt
Bioinformatics 2018, 34 (3): 530-532
Online | ETH Reserach Collection | Project page
 |
Mahito Sugiyama, Karsten BorgwardtHalting in Random Walk Kernels |
Code
A fast C++ implementation of graph kernels is available at our GitHub repository here, which includes:
- simple kernels between vertex and/or edge label histograms
- random walk kernels (popular baselines)
- Weisfeiler-Lehman graph kernel (state-of-the-art)
Datasets
The GraphML format of the graph datasets MUTAG, NCI1, NCI109, ENZYMES, and D&D is also available Download here (ZIP, 12 MB).
Publication
Halting in Random Walk Kernels
Mahito Sugiyama and Karsten Borgwardt
Advances in Neural Information Processing Systems 28 (NIPS 2015), 1639-1647
Online | ETH Research Collection | Project page | GitHub
Contact Karsten Borgwardt for questions.
 |
|
Code and Datasets
The files are available at our GitHub repository here. They include
- a MATLAB implementation of the GraphHopper kernel as proposed in [1],
- an implementation of the propagation kernel as presented in [2], and
- the datasets used in the papers.
Publications
[1]
Scalable kernels for graphs with continuous attributes
Aasa Feragen, Niklas Kasenburg, Jens Petersen, Marleen de Bruijne and Karsten Borgwardt
Advances in Neural Information Processing Systems 26 (NIPS 2013), 216-224
Online | ETH Research Collection | Project page | GitHub
[2]
Efficient graph kernels by randomization
Marion Neumann, Novi Patricia, Roman Garnett and Kristian Kersting
Lecture Notes in Computer Science 7523 "Machine Learning and Knowledge Discovery in Databases. ECML PKDD 2012.", 378–393
Online
 |
|
Code
This zip (ZIP, 580 KB) archive contains Matlab scripts to compute various graph kernels for graphs with unlabeled or categorically labeled nodes, such as the random walk, shortest path, graphlet, several instances of Weisfeiler-Lehman or other subtree kernels. For a more detailed list of available kernels please consult the README (3 KB) in the archive.
Datasets
Download the graph data sets MUTAG, NCI1, NCI109, ENZYMES, and D&D (all in Matlab format) as a zip (ZIP, 10.1 MB) or a tar.gz (GZ, 10.1 MB) archive. The archive also contains a README describing the data sets and explaining their usage. Download the Matlab scripts for the WL subtree, edge and shortest path kernels as a zip (ZIP, 8 KB) or a tar.gz (GZ, 4 KB) archive. This archive also contains a README explaining the usage of the scripts.
Publication
Weisfeiler-Lehman Graph Kernels
Nino Shervashidze, Pascal Schweitzer, Erik Jan van Leeuwen, Kurt Mehlhorn and Karsten M. Borgwardt
Journal of Machine Learning Research (JMLR) 2011, 12: 2539−2561
Online | ETH Research Collection | Project page including code
Contact Karsten Borgwardt for questions.
 |
|
Code
The Matlab code can be found here (M, 8 KB).
Datasets
Download datasets in Matlab format (Data (MAT, 11.4 MB)).
Publication
Graph Kernels
SVN Vishwanathan, Nic Schraudolph, Risi Kondor and Karsten M. Borgwardt
Journal of Machine Learning Research (JMLR) 2010, 11: 1201−1242
Online | ETH Research Collection | Project page including code
Contact Karsten Borgwardt for questions.
Introduction
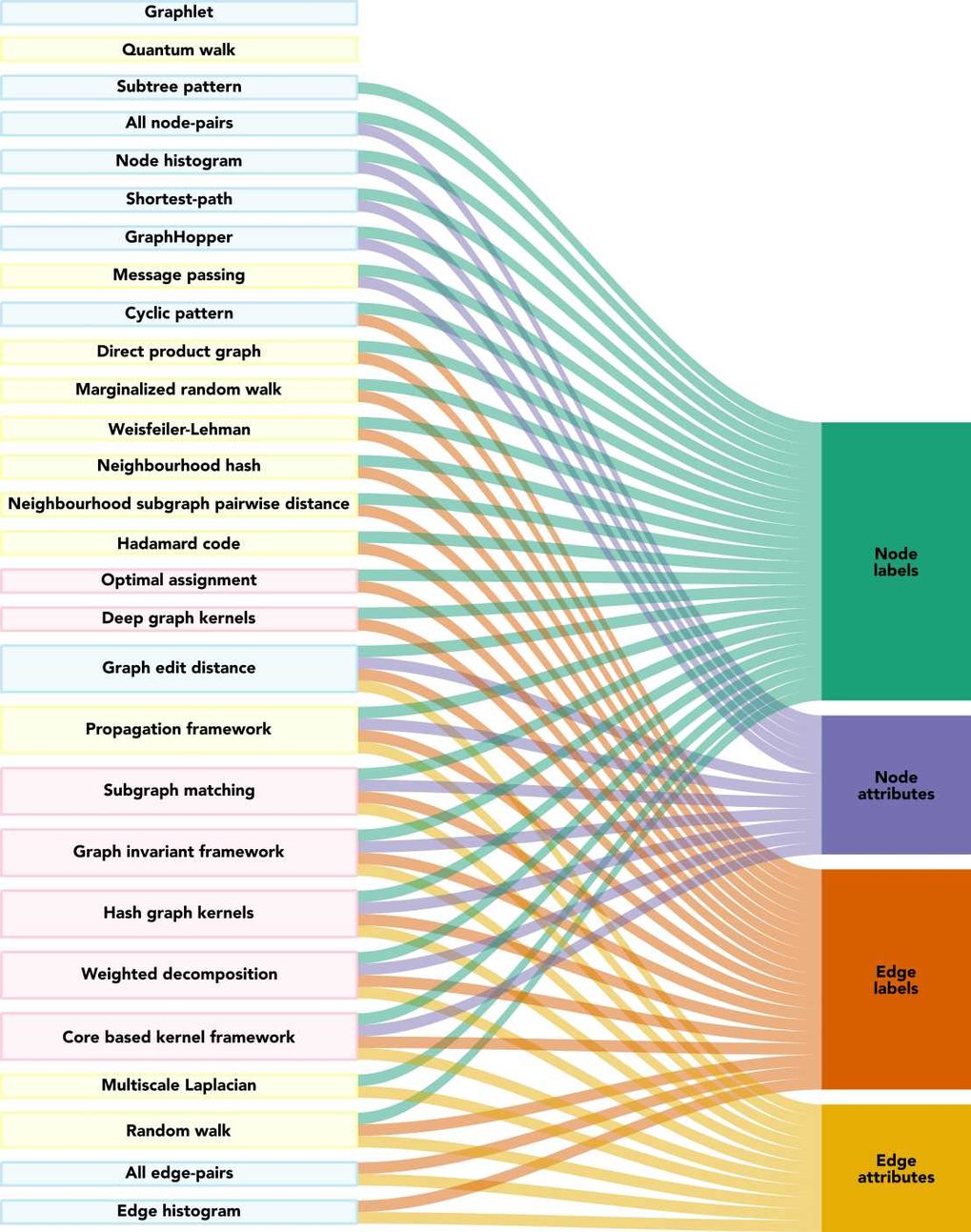
Graph-structured data are an integral part of many application domains, including chemoinformatics, computational biology, neuroimaging, and social network analysis. Over the last two decades, numerous graph kernels, i.e. kernel functions between graphs, have been proposed to solve the problem of assessing the similarity between graphs, thereby making it possible to perform predictions in both classification and regression settings. Our review covers existing graph kernels, their applications, software plus data resources, and an empirical comparison of state-of-the-art graph kernels.
Code
The code used for our experiments is available on GitHub.
Dataset
Our paper used datasets from the TUDortmund benchmark datasets for graphs repository. The repository has since been migrated to www.graphlearning.io.
Figures
The visualizations of the graph kernels used in the paper are available as a PDF here (PDF, 2.8 MB).
Contributors
Karsten Borgwardt
Elisabetta Ghisu (alumna)
Felipe Llinares-López (alumnus)
Leslie O'Bray
Bastian Rieck (alumnus)
References
Graph Kernels: State-of-the-Art and Future Challenges
Karsten Borgwardt, Elisabetta Ghisu, Felipe Llinares-López, Leslie O'Bray, and Bastian Rieck
Foundations and Trends® in Machine Learning 2020, 13 (5-6): 531-712.
Online | Project page | arXiv