Feeling the need for speed? 100-times faster DNA-PAINT
Scientists at the Max Planck Institute (MPI) of Biochemistry enable 100-times faster multiplexed DNA-PAINT microscopy using optimized DNA sequences
• DNA-PAINT uses DNA-barcoded probes to visualize nanoscale biological structures
• Optimized DNA designs enable 100-times faster and multicolor imaging
• High throughput and molecular resolution microscopy might in the future improve our understanding of the interactions between different tumor markers
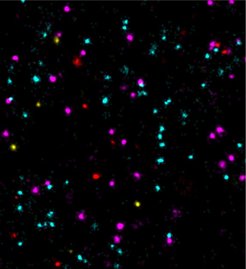
Super-resolution fluorescence microscopy can be used to visualize structures smaller than 200 nanometers, i.e. below the diffraction limit of light. One of the microscopy techniques, called DNA-PAINT, was developed by Ralf Jungmann, research group leader at the MPI of Biochemistry and Professor for Experimental Physics at the Ludwig Maximilian University Munich together with colleagues. The technique uses short ‘imagers’, dye-labeled DNA strands that temporarily bind to their target molecules in a complementary manner to produce the necessary "blinking" for super-resolution reconstruction of the images.
“We have recently improved DNA-PAINT’s traditionally rather slow acquisition speed by an order of magnitude by optimizing DNA sequence design.” says Jungmann. "However, this came at the cost of losing multiplexing, which means that several structures in the cell cannot be observed simultaneously", added Jungmann. The simultaneous observation of several proteins, however, is important for the better understanding of complex signaling cascades between tumor and normal cells.
This multiplexing capability was not achievable in speed-optimized DNA-PAINT, as only a single optimized sequence with improved hybridization characteristics was available. “We asked ourselves how to allow multiplexed imaging and, at the same time, push image acquisition speed even further”, says Sebastian Strauss, first author of the paper and co-worker in Jungmann’s group.
In the current study, the researchers present a novel concept that has successfully improved the imaging speed. They took advantage of the fact that the frequency of binding of the imagers to their target strands scales linearly with the number of available binding sites. "The more binding sites there are, the faster the image acquisition proceeds. However, simply concatenating binding sites would lead to undesirably long docking sequences, potentially reducing achievable image resolution and increasing non-specific binding," says Strauss. To circumvent these issues, the researchers designed repetitive sequence motifs, e.g. (TCC)n, which could be concatenated to provide overlapping binding sites yet only slightly increasing strand length. "We designed six individual, periodic sequence motifs, which allows us to introduce multiplexing to speed-optimized DNA-PAINT," said Strauss. "In combination with previous improvements, we can now speed up DNA-PAINT by a factor of 100," adds Jungmann.
From the breadboard to cell applications
To optimize the new sequence motifs and benchmark their improvements, the group used DNA origami structures, which are self-assembled, nanometer-sized DNA objects autonomously folding into predefined shapes. These structures can be used to arrange DNA-PAINT binding sites spaced precisely at e.g. 5-nm distances. This enabled the researchers to evaluate the improvements to DNA-PAINT under defined conditions. “The new optimized DNA sequences allowed us to resolve six different DNA origami structures instead of only one, in just a few minutes”, explains Strauss.
“We are thrilled to apply the now further improved imaging speed in DNA-PAINT to tackle biological questions. For instance, tumor markers could previously only be examined slowly and not clearly on a single-molecule level. In our study, the measurement of four different tumor markers confirms a fast and accurate analysis of their molecular positions and interactions. This could provide important insights for drug development and their mechanisms of action," concludes Jungmann.
Original Publication:
S. Strauss & R. Jungmann, Nature Methods, June 2020
DOI: https://doi.org/10.1038/s41592-020-0869-x
