DNA-PAINT super-resolution microscopy at speed
Optimized DNA sequences allow for 10-times faster image acqusition in DNA-PAINT
• DNA-PAINT uses DNA-based probes to visualize tiny cellular structures
• The technique has thus far been limited by rather slow image acquisition speeds
• Optimized DNA sequence design now yields imaging results 10-times faster, enabling high-throughput studies with biomedical relevance
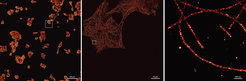
Recent advances in fluorescence microscopy allow researchers to study biological processes below the classical diffraction limit of light. Ralf Jungmann, research group leader at the Max Planck Institute of Biochemistry and Professor for Experimental Physics at the University of Munich, and colleagues developed DNA-PAINT, a variant of these so-called super-resolution approaches. “DNA-PAINT yields super-resolved images using comparably simple microscopes”, says Jungmann. The technique uses short, dye-labeled DNA strands that transiently interact with their target-bound complements in order to create the necessary “blinking” for super-resolution reconstruction. This approach enables sub-10-nm spatial resolution and easy multiplexing through the use of orthogonal DNA sequences for different targets.

“During the last years, we have optimized DNA-PAINT in a few key areas. However, one major limitation still persists, which prevents DNA-PAINT to be applied to biomedically relevant high-throughput studies: The rather slow image acquisition speed”, says Jungmann. Classical DNA-PAINT experiments can easily last from tens of minutes to hours. “We have checked carefully why this takes so long”, says Florian Schüder, lead author of the current study and co-worker in Jungmann’s group. “Optimized DNA sequence design and improved image buffer conditions allowed us to speed things up by an order of magnitude”, adds Schüder. To achieve this, the researchers teamed up with the Department of Petra Schwille at the MPIB to investigate the influence of DNA sequence and base composition on hybridization speed (or the formation of the double helix). The strands used in DNA-PAINT generally consist of all four DNA bases: Adenine (A), Thymine (T), Guanine (G), and Cytosine (C). It is generally well understood how base composition and strand length modulate the binding time of the complementary strands: The longer the strands and the higher their GC-content, the more stable the duplex becomes, resulting in an increased binding time. However, the influence on the hybridization rate (and thus the DNA-PAINT image acquisition speed), specifically for short oligonucleotides, is poorly understood. In the current study, the researchers could show that the formation of intramolecular hairpin motifs (e.g. strands folding back on themselves), present in even short oligonucleotides, could be completely avoided by limiting the bases used in the sequence design to only two out of the four (e.g. T and C instead of A, T, C, or G). “We are now designing strands using only a so-called two-letter alphabet (e.g. only using T and C or A and G)”, says Schüder. “In combination with additional optimizations ot the imaging buffer, we could squeeze out another factor of two in speed, which now allows us to acquire images 10-times faster than before”, adds Schüder.
From the DNA origami breadboard to cells
In order to quantitatively assess the improvements to DNA-PAINT, the researchers used DNA origami structures, which are self-assembled, nanometer-sized DNA objects autonomously folding into predefined shapes. These structures can be used to arrange DNA-PAINT binding sites spaced precisely at e.g. 5-nm distances. This allowed the researchers to evaluate the speed improvement in DNA-PAINT using well-defined conditions. In a next step, the team applied the speed improvement also to a cellular system. For this, microtubules, which are part of the cytoskeleton, were visualized at super-resolution, 10-times faster than before. “The increased imaging speed allowed us to acquire an area of one square millimeter at a resolution of 20 nm in only 8 hours. This would have taken us almost four days before”, explains Schüder.
Ralf Jungmann concludes: “With these current improvements, which allow us to image 10-times faster, we bring DNA-PAINT to the next level. It should now be feasible to apply it to high-througphut studies with biological and biomedical relevance e.g. in diagnostic applications.”
Original Publication
F. Schüder, J. Stein, F. Stehr, A. Auer, B. Sperl, M.T. Strauss, P. Schwille und R. Jungmann: An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions. Nature Methods, Oktober 2019.
DOI: https://doi.org/10.1038/s41592-019-0584-7

