Chemotaxis and phototaxis of Halobacterium salinarum
I. Introduction
Halobacterium salinarum is a motile halophilic archaeon that is able to sense many external stimuli and reacts by actively searching for the best place to live. A signal transduction cascade is involved in this chemo- and phototactic response. Signals are sensed by specific receptors, amplified and integrated by a two-component regulatory system, and finally affect the flagellar motor as target. This results in a "biased random walk" which permits Halobacterium to swim into the desired direction.
Here we provide an introduction to various aspects of this complex physiological response (motility of Halobacterium, physiological responses, modules and molecules of the cascade, the flagellar apparatus, modelling, and analysis techniques) and provide links to more detailed descriptions.
Detailed and quantitative experimental data for the phototactic response of Halobacterium have been obtained. The combination of detailed experimentation and modelling makes this response a very useful model case for a systems biological approach.
II. Motility of Halobacterium salinarum

The archaeal flagella
Halobacterium moves by means of a polarly inserted flagellar bundle which consists of 5 to 10 flagella. The archaeal motor functions by a rotatory principle, resulting in rotation of the attached flagella. This is the same principle applied by their bacterial counterparts, but in contrast its architecture is completely different.
The mode of action of the motor is based on a set of 10-15 filaments that form a right-handed helical bundle which, if not stimulated, rotates in an unbiased way. (Alam and Oesterhelt 1984). The motor spends equal times in the clockwise (forward swimming) and counterclockwise (reverse swimming) modes and the two modes are separated by stops of about 100 ms length. The archaeal flagella do not disentangle upon reversal (Alam and Oesterhelt 1984), in contrast to their eubacterial counterparts.
References from the department:
- M. Alam, D. Oesterhelt:Morphology, Function and Isolation of Halobacterial Flagella. J. Mol. Biol. 176, 459-475 (1984)
- J. Kupper, W. Marwan, D. Typke, H. Grünberg, U. Uwer, M. Gluch, D. Oesterhelt:The Flagellar Bundle of Halobacterium salinarium is Inserted into a Distinct Polar Cap Structure. J. Bacteriol. 176, 5184-5187 (1994).
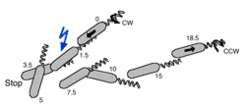
Directional moving by a "biased random walk"
In the absence of any stimulus, H. salinarum performs a random walk based on an unbiased alternation between clockwise to counterclockwise rotation of the flagellar bundle. Stimuli affect the timing of the motor reversal by activation or inhibition of the flagellar motor switch. If negative stimuli such as blue light or repellent chemicals are applied the motor reversal occurs premature. As a consequence, the organism swims into the opposite direction and thus moves away from the repellent stimulus.
Increasing attractant stimuli inhibit the switch of the flagellar motor, resulting in prolonged swimming into the current direction. As a consequence, the organism will get closer to the attractant.
The bias introduced by a stimulus causes accumulation or decumulation of cells depending on its nature in areas of enhanced stimulus concentration.
The following fundamental characteristics of archaeal taxis should be noted:
-
No distinction is made between negative stimuli, which may be either caused by increasing repellent or by decreasing attractant signals (also not for the two types of positive stimuli).
-
Stimuli have the same effect independent of the mode of swimming (forward or reverse), i.e. independent of the current direction of flagellar rotation.
III. Physiological responses
Introduction
Halobacterium can sense a large number of stimuli as revealed by physiological experiments which allowed to identify 8 different signals. The cells have capabilities for phototaxis (three colors), for chemotaxis (towards nutrients and compatible osmolytes), for aerotaxis (oxygen), and for sensing of its membrane potential.
An additional 10 transducers with yet unassigned function have been identified in the genome by homology searches and 2 of these most likely are also involved in chemotaxis as revealed by gene neighbourhood analysis. Thus, Halobacteriumshows a highly complex perception of its environment.
Phototaxis
Halobacterium salinarum lives under bright tropical sunshine (where damaging UV light intensity is high) and carries out photosynthesis with the help of two light-driven ion pumps, acteriorhodopsin and alorhodopsin, which - as a first and direct effect - energize the membrane.
The organism developed a strategy to find optimal conditions with the help of three receptors.
One receptor, MpcT, functions at high light intensity and is a sensor of the membrane potential changes (Koch and Oesterhelt 2005), which result from variation in the activity of bacteriorhodopsin as a consequence of changing light intensity (irradiance) (Bibikov et al. 1993, Bibikov et al. 1991, Grishanin et al. 1996). At dropping membrane potential, MpcT will make the cell stop and swim back.
At low light intensities, the two photoreceptors SRI and SRII (sensory rhodopsins I and II) can sense three light colors and help to find valuable orange light where photosynthesis is optimal, but to avoid damaging UV or useless blue light. SRI detects two colors: orange light and - in the presence of orange background light - UV light. SRII detects blue light.
At high light intensity the two colour sensors SRI and SRII are blind because they become photochemically bleached and thus phototaxis depends on MpcT.
Chemotaxis (including osmotaxis and aerotaxis)
Two amino acid receptors have been characterized for Halobacterium: Car (cytoplasmic arginine sensor) (Storch et al. 1999) and BasB/BasT (branched and sulfur-containing amino acid sensor) (Kokoeva and Osterhelt 2000, Kokoeva et al. 2002). In addition, a sensor for compatible osmolytes (CosB/CosT) has been characterized (Kokoeva et al. 2002).
As a microaerobic organism, Halobacterium searches for an intermediate oxygen concentration. An aerophilic receptor (HtrVIII, responsible for cell accumulation around an air bubble) and an aerophobic receptor (HemAT, Hou et al. 2000) have been identified.
IV. Modules and molecules of the signal transduction cascade
Introduction
We describe 4 modules of the signal transduction cascade.
-
Signal reception:
The first module is involved in reception of the signal. The various stimuli are detected by specific receptors (or receptor domains). With the help of transducers, the receptors trigger intracellular signaling pathways. All of the 18 Halobacterium transducers report to a single histidine kinase, CheA. -
Signal integration and amplification:
The second module is the two-component regulatory system consisting of the histidine kinase CheA and the response regulator CheY. This module integrates and amplifies the signal. The output of this module is phosphorylated CheY, the concentration of which determines the switching frequency of the flagellar motor. -
Targetting to the motility system:
The third module is the flagellar motor and its switch, which is the target of this signal transduction cascade and permits movement into the desired direction. -
Adaptation:
The fourth module is the adaptation module which involves covalent modification of the transducers by methylation of glutamate residues. The level of methylation results from the activity of two antagonistic enzymes, the constitutive methyltransferase CheR and a regulated methylesterase CheB.
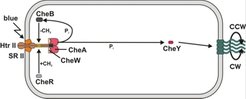
Signal reception (receptors/transducers)
Signals are received by receptor molecules or domains which are as diverse as the signals themselves. Transducers transfer the signal via their highly conserved signalling domain to the two-component regulatory system through activation of the histidine kinase CheA. The conservation of the signalling domain permits reliable identification of transducers by simple homology searches.
The receptor may be an independent protein which forms a complex with its transducer (SRI/HtrI, SRII/HtrII, BasB/BasT, CosB/CosT). Alternatively, the receptor and transducer domains may be fused into a single polypeptide chain (HtrVIII).
Many signals are received outside of the cell and correspondingly the receptors are membrane proteins (SRI, SRII, HtrVIII, MpcT) or extracellular (periplasmic) proteins (BasB, CosB). Other receptors/transducers are soluble proteins which detect signals within the cell (Car, HemAT).
In toto, Halobacterium has 18 distinct transducers as revealed by genome analysis. Of these, 8 have been functionally characterized. The gene context of another 2 transducers makes it very likely that they participate in chemotaxis.
Here is a brief list of functionally characterized receptors/transducers:
involved in phototaxis: (MpcT, SRI/HtrI, SRII/HtrII)
- involved in chemotaxis (and osmotaxis): (Car, BasB/BasT, CosB/CosT)
- oxygen sensing: (HemAT, HtrVIII)
Transducers are subject to methylation of glutamate residues in the adaptation response. Therefore, they are also called methyl-accepting chemotactic proteins (MCP). Methylated transducers are more active than demethylated transducers.
References from the department:
- M.K. Koch, D. Oesterhelt:MpcT is the transducer for membrane potential changes in Halobacterium salinarum. Mol. Microbiol. 55, 1681-1694 (2005).
- M.V. Kokoeva, D. Oesterhelt:BasT, a Membrane-Bound Transducer Protein for Amino Acid Detection in Halobacterium salinarum. Mol. Microbiol. 35, 647-656 (2000).
- M.V. Kokoeva, K.-F. Storch, Ch. Klein, D. Oesterhelt:A Novel Mode of Sensory Transduction in Archaea: Binding Protein Mediated Chemotaxis towards Osmoprotectrants and Amino Acids in Halobacterium salinarum. EMBO J. 21, 2312-2322 (2002).
- M.Krah, W.Marwan, A. Vermeglio, D. Oesterhelt:Phototaxis of Halobacterium salinarium Requires a Signalling Complex of Sensory Rhodopsin I and its Methyl-accepting Transducer HtrI. EMBO J. 13, 2150-2155 (1994).
- J. Rudolph, B. Nordmann, K.-F. Storch, H. Gruenberg, K. Rodewald, D. Oesterhelt:A Family of Halobacterial Transducer Proteins. FEMS Microbiology Lett. 139, 161-168 (1996).
- K.-F. Storch, J. Rudolph, D. Oesterhelt:Car: A Cytoplasmic Sensor Responsible for Arginine Chemotaxis in the Archaeon Halobacterium salinarum. EMBO J. 18, 1146-1158 (1999).
The two-component regulatory system (CheA and CheY)
The two-component regulatory system, which resembles their eubacterial counterpart, consists of the histidine kinase CheA and the response regulator CheY (Rudolph et al. 1995). This module integrates various signals (as all transducers signal to the sole histidine kinase CheA pool) and also amplifies the signal (due to the involvement of catalytic activities).
Upon activation of CheA, a phosphate group is first attached by autophosphorylation to a histidine residue of CheA. The catalytic residue is a distinct histidine on the other chain of the homodimeric enzyme (i.e. the autocatalytic reaction is in trans). The phosphate is then transferred to an aspartate residue of the response regulator CheY, a small and highly conserved protein. The concentration of phosphorylated CheY is the output of this module and determines the switching frequency of the flagellar motor.
CheY is regenerated in its non-phosphorylated form by a yet uncharacterized hydrolytic reaction.
It should be noted that the phosphate cannot only be transferred from CheA to CheY but also to other proteins having a cheY domain, most notably the methylesterase CheB, which is involved in adaptation.
References from the department:
- J. Rudolph, D. Oesterhelt:Chemo- and Phototaxis Require a CheA Histidine Kinase in the Archaeon Halobacterium salinarium. EMBO J. 14, 667-673 (1995)
- J. Rudolph, N. Tolliday, C. Schmitt, S. Schuster, D. Oesterhelt:Phosphorylation in Halobacterial Signal Transduction. EMBO J. 14, 4249-4257 (1995)
- J. Rudolph, D. Oesterhelt:Deletion Analysis of the che Operon in the Archaeon Halobacterium salinarium. J. Mol. Biol. 258, 548-554 (1996)
The target: the flagellar motor and its switch
Directed swimming of Halobacterium originates from introduction of a bias into its random walk. This bias results from regulation of the switch frequency of the flagellar motor, which is the target of the signal transduction cascade.
In all archaea, the flagellum itself is a heteromultimer of flagellins. In Halobacteriumthere are five distinct flagellins (FlgA1, FlgA2, FlgB1, FlgB2, FlgB3). Biogenesis of the flagella depends on genes in the fla cluster which is located between the che and flgB genes.
Although the flagellar motor and its switch have been characterized physiologically in detail, none of their components have yet been identified by genetic or biochemical analysis.
More details on the flagella, flagellar motor, and flagellar biogenesis are described in a later section.
The adaptation system (CheB and CheR)
Adaptation, which is required to measure changes in signal strength rather than their absolute level, is considered to be a distinct module which interacts with the reception and integration/amplification modules. It consists of two antagonistic enzymes which methylate (CheR) and demethylate (CheB) glutamate residues in well-conserved methylation domains on the transducers.
The activity of the methyltransferase CheR is constitutive while the activity of the demethylase CheB is regulated by phosphorylation (phosphorylated CheB is active). CheB contains a cheY domain which may receive phosphate groups from the phosphorylated histidine kinase CheA. Thus, phosphorylation of CheA does not only activate the switch of the flagellar motor but also activates the adaptation response (negative feedback).
Additional chemotactic proteins
The chemotaxis gene cluster in Halobacterium contains additional genes, some of which have been shown to be involved in the signal transduction cascade and to affect chemo- and phototaxis.
- CheC:
Deletion of one of the three CheC genes resulted in introduction of a bias towards forward swimming of Halobacterium(Rudolph and Oesterhelt 1996). - CheW:
CheW is a coupling protein which is involved in the interaction between transducers and the histidine kinase CheA. Halobacterium has two isoforms of this protein, CheW1 and CheW2. - CheD:
The molecular function of CheD is not yet known.
V. The flagella and their biogenesis
The flagellar motor and its switch
None of the molecular components of the archaeal motor has so far been identified neither from genomic nor from biochemical analysis. Homology searches with components of the eubacterial motor are not successful as the molecular architecture of the two motors seems to be completely different.
Despite the lack of molecular data, the motor could be functionally characterized in detail due to a detailed quantitative analysis of the phototactic response followed by in-depth modelling of the motor's switch behaviour.
Note:
In addition to phosphorylated CheY, the metabolite fumarate plays an important role in the flagellar motor switch events (Marwan et al. 1990, Montrone et al. 1998).
The flagellum and the flagellins
The molecular architecture of the archaeal flagellum is well known and is principally different from that of eubacterial flagella. The archaeal flagellum forms a right-handed helix and is a polymer of several flagellins, which are homologous to each other. Flagellins are glycoproteins.
Halobacterium produces five distint flagellins, FlgA1, FlgA2, FlgB1, FlgB2, and FlgB3.
Flagellins are synthesized as precursors. The presequence is cleaved off by preflagellin peptidase (flaK). Flagellins are exported through a specific export system consisting of FlaI and FlaJ.
References from the department:
- N. Patenge, A. Berendes, H. Engelhard, S.C. Schuster, D. Oesterhelt:The fla Gene Cluster is Involved in the Biogenesis of Flagella in Halobacterium salinarum. Mol. Microbiol. 41, 1-12 (2001).
The motility and signal transduction cluster
A cluster of fla genes is found adjacent to flagellin genes and occurs throughout the kingdom with a high variability of gene arrangements and several protein and domain duplications.
The genes flaI and flaJ most likely form the flagellin export system.
The molecular function of the other flagellin genes (FlaD, FlaCE, FlaF, FlaG, FlaH) has not yet been characterized in detail.
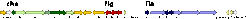
VI. Comparison of archaeal and eubacterial chemotaxis and motility genes
The genetics and the detailed biological mechanisms by which taxis is controlled are best understood in the eubacterium Escherichia coli. All components of the chemotaxis pathway of E. coli have been identified and they are very well characterized. In addition, chemotaxis in Bacillus subtilis is well studied and shows a number of remarkable differences to E. coli.
Several chemotactic proteins from eubacteria and archaea are clearly homologous and are most likely isofunctional:
-
transducers with their highly conserved signalling domain
-
histidine kinase CheA
-
response regulator CheY
-
methyltransferase CheR
-
methylesterase CheB
-
coupling protein CheW
-
cheZ is present in E.coli but missing in B.subtilis and in archaea
-
cheD is present in B.subtilis and in archaea but missing in E.coli
-
cheC is present in B.subtilis and in archaea but missing in E.coli
Eubacteria contain a number of periplasmic substrate-binding proteins which interact in ABC-type transport systems. In E.coli, the dual function of substrate-binding proteins in both, transport and chemotactic recognition, could be shown. Dual-function substrate-binding proteins are encoded in transcription units with the corresponding ABC-type transport system and not with the functionally associated transducer.
In Halobacterium, four transducer genes are adjacent to genes which are homologous to eubacterial periplasmic substrate-binding proteins and functional characterization of two of the gene pairs (basB/basT and cosB/cosT) proved that they are involved in chemotaxis/osmotaxis.
Although the molecules involved in the signal transduction cascade are clearly homologous, the flagellar apparatus of eubacteria and archaea has a completely different architecture. Eubacterial flagella are hollow fibers and new flagellin subunits are transported through the internal channel and then incorporated at the tip. Archaeal flagella contain glycosylated flagellin subunits which are most likely incorporated at the base and their internal channel is not wide enough for flagellin export. While eubacterial flagella disentangle upon reversal of the flagellar rotation, archaeal flagella do not change their structure and propell the cell in both modes of rotation.
The flagellar motor must also have a completely distinct architecture. Although the eubacterial motor is known in detail, components of the archaeal motor could not be identified by homology searches in any of the sequenced genomes.
VII. Modelling of the phototactic response
The phototactic response has been modelled in detail (Nutsch et al. 2003, Nutsch et al., 2005).
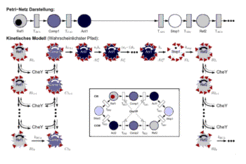
References from the department:
- T. Nutsch, D. Oesterhelt, E.D. Gilles and W. Marwan:
The Switch Cycle of an Archaeal Flagellar Motor and its Sensory Control.
Biophys. J. 89, 2307-2323 (2005). - T. Nutsch, W. Marwan, D. Oesterhelt, E.-D. Gilles:
Signal Processing and Flagellar Motor Switching during Phototaxis of Halobacterium salinarum.
Genome Res. 13, 2406-2412 (2003).
VIII. Analysis techniques
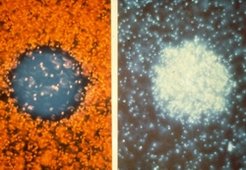
Swarm plates
Cells are stabbed into soft agar plates which allows them to move. Growing cells form concentric rings if they are motile and chemotactically active.
Capillary assays
Capillaries filled with chemotactically active substances are inserted into cell suspensions. Cells will swim randomly into the capillaries but may accumulate when these are filled with attractants. After the experiment, the number of cells in the capillaries is quantified.
Motion analysis
Cells are visualized under the microscope and their movements are recorded by a video system. Cell tracking softward allows to analyze the cell's swimming behaviour. Light stimulation in this setup permits a quantitative analysis of the phototactic response.
For further details, see motion analysis of Halobacterium salinarum
Flow assays for detection of transducer demethylation
Adaptation is due to covalent methylation and demethylation of transducers. Upon demethylation, free methanol is released. If the methyl groups on the transducers are labelled (by addition of labelled methionine), the released methanol is also labelled and thus the rate of demethylation can be determined in a flow assay.
(for H. salinarum described in Nordmann et al. 1994)
Functional analysis by gene deletion
Molecular biological techniques can be used to eliminate genes from an organisms. The phenotypic effects of such mutants can be studied in detail.
Related web pages from the department
-
phototaxis as model case for a systems biological approach
Subject-related publications from the department
- T. Nutsch, D. Oesterhelt, E.D. Gilles and W. Marwan:
The Switch Cycle of an Archaeal Flagellar Motor and its Sensory Control.
Biophys. J. 89, 2307-2323 (2005). - M.K. Koch, D. Oesterhelt:
MpcT is the transducer for membrane potential changes in Halobacterium salinarum.
Mol. Microbiol. 55, 1681-1694 (2005). - T. Nutsch, W. Marwan, D. Oesterhelt, E.-D. Gilles:
Signal Processing and Flagellar Motor Switching during Phototaxis of Halobacterium salinarum.
Genome Res. 13, 2406-2412 (2003) - M.V. Kokoeva, K.-F. Storch, Ch. Klein, D. Oesterhelt:
A Novel Mode of Sensory Transduction in Archaea: Binding Protein Mediated Chemotaxis towards Osmoprotectrants and Amino Acids in Halobacterium salinarum.
EMBO J. 21, 2312-2322 (2002). - N. Patenge, A. Berendes, H. Engelhard, S.C. Schuster, D. Oesterhelt:
The fla Gene Cluster is Involved in the Biogenesis of Flagella in Halobacterium salinarum.
Mol. Microbiol. 41, 1-12 (2001). - M.V. Kokoeva, D. Oesterhelt:
BasT, a Membrane-Bound Transducer Protein for Amino Acid Detection in Halobacterium salinarum.
Mol. Microbiol. 35, 647-656 (2000). - [toadd] W. Marwan, D. Oesterhelt:
Archaeal Vision and Bacterial Smelling: Sensory Control of the Swimming Behaviour by Two Component Signalling and Fumarate.
ASM-News 66, 83-90 (2000). - [toadd] W. Marwan, D. Oesterhelt:
Die Phototaxis der Halobakterien: Biochemische Mechanismen einer einfachen Verhaltensreaktion.
Chemie in unserer Zeit 33, 141-151 (1999). - M. Montrone, M. Eisenbach, D. Oesterhelt, W. Marwan:
Regulation of Switching Frequency and Bias of the Bacterial Flagellar Motor by CheY and Fumarate.
J. Bacteriol. 180, 3375-3380 (1998). - Montrone, M., D. Oesterhelt and W. Marwan:
Metabolic Phosphorylation-Independent Bacterial Chemoresponses Correlate with Changes in the Cytoplasmic Level of Fumarate.
J. Bacteriol. 178, 6882-6887 (1996). - J. Rudolph, B. Nordmann, K.-F. Storch, H. Gruenberg, K. Rodewald, D. Oesterhelt:
A Family of Halobacterial Transducer Proteins.
FEMS Microbiology Lett. 139, 161-168 (1996). - J. Rudolph, D. Oesterhelt:
Deletion Analysis of the che Operon in the Archaeon Halobacterium salinarium.
J. Mol. Biol. 258, 548-554 (1996). - U. Haupts, E. Bamberg. D. Oesterhelt:
Different Modes of Proton Translocation by Sensory Rhodopsin I.
EMBO J. 15, 1834-1841 (1996). - D. Oesterhelt, W. Marwan:
Wie Archaea sehen und schmecken.
Biospektrum 1, 11-16 (1995). - K.-F. Storch, J. Rudolph, D. Oesterhelt:
Car: A Cytoplasmic Sensor Responsible for Arginine Chemotaxis in the Archaeon Halobacterium salinarum.
EMBO J. 18, 1146-1158 (1999). - J. Rudolph, N. Tolliday, C. Schmitt, S. Schuster, D. Oesterhelt:
Phosphorylation in Halobacterial Signal Transduction.
EMBO J. 14, 4249-4257 (1995). - U. Haupts, Ch. Haupts, D. Oesterhelt:
The Photoreceptor Sensory Rhodopsin I as a Two-photon-driven Pump.
Proc. Natl. Acad. Sci. USA, 92, 3834-38838 (1995). - J. Rudolph, D. Oesterhelt:
Chemo- and Phototaxis Require a CheA Histidine Kinase in the Archaeon Halobacterium salinarium.
EMBO J. 14, 667-673 (1995). - U. Haupts, W. Eisfeld, M. Stockburger, D. Oesterhelt:
Sensory rhodopsin I Photocycle Intermediate SRI 380 Contains 13-cis Retinal Bound via an Unprotonated Schiff Base.
FEBS Lett. 356, 25-29-(1994). - W. Marwan, S.I. Bibikov, M. Montrone, D. Oesterhelt:
Mechanism of Photosensory Adaptation in Halobacterium salinarium.
J. Mol. Biol. 246, 493-499 (1995). - R. Seidel, B. Scharf, M. Gautel, K. Kleine, D. Oesterhelt, M. Engelhard:
The Primary Structure of Sensory Rhodopsin II: The First Member of a New Retinal Protein Subgroup is Coexpressed with its Transducer, the Halobacterial Transducer of Rhodopsin II.
Proc. Natl. Acad. Sci. 92, 3036-3040 (1995). - M. Krah, W. Marwan, D. Oesterhelt:
A Cytoplasmic Domain is Required for the Functional Interaction of SRI and HtrI in Archaeal Signal Transduction.
FEBS Lett. 353, 301-304 (1994). - J. Kupper, W. Marwan, D. Typke, H. Grünberg, U. Uwer, M. Gluch, D. Oesterhelt:
The Flagellar Bundle of Halobacterium salinarium is Inserted into a Distinct Polar Cap Structure.
J. Bacteriol. 176, 5184-5187 (1994). - B. Nordmann, M. R. Lebert, M. Alam, S. Nitz, H. Kollmannsberger, D. Oesterhelt, G. L. Hazelbauer:
Identification of Volatile Forms of Methyl Groups Released by Halobacterium salinarium .
J. Biol. Chem. 269, 16449-16454 (1994). - M.Krah, W.Marwan, A. Vermeglio, D. Oesterhelt:
Phototaxis of Halobacterium salinarium Requires a Signalling Complex of Sensory Rhodopsin I and its Methyl-accepting Transducer HtrI.
EMBO J. 13, 2150-2155 (1994). - M. Montrone, W. Marwan, H. Grünberg, S. Mußeleck, Ch. Starostzik, D. Oesterhelt:
Sensory Rhodopsin-Controlled Release of the Switch Factor Fumarate in Halobacterium salinarium.
Mol. Microbiol. 10(5), 1077-1085 (1993). - S. Bibikov, R. Grishanin, A. Kaulen, W. Marwan, D. Oesterhelt, V. Skulachev:
Bacteriorhodopsin is Involved in Halobacterial Photoreception.
Proc. Natl. Acad. Sci. USA. 90, 9446-9450 (1993) - E. Ferrando-May, B. Brustmann, D. Oesterhelt:
A C-terminal Truncation Results in High-level Expression of the Functional Photoreceptor Sensory Rhodopsin I in the Archaeon Halobacterium salinarium.
Mol. Microbiol. 9, 943-953 (1993). - E. Ferrando-May, M. Krah, W. Marwan, D. Oesterhelt:
The Methyl-accepting Transducer Protein HtrI is Functionally Associated with the Photoreceptor Sensory Rhodopsin I in the Archaeon Halobacterium salinarium.
EMBO J. 12, 2999-3005 (1993). - S. Bibikov, R. Grishanin, W. Marwan, D. Oesterhelt, V. Skulachev:
The Proton Pump Bacteriorhodopsin is a Photoreceptor for Signal Transduction in Halobacterium halobium.
FEBS Lett. 295, 223-226 (1991). - W. Marwan, D. Oesterhelt:
Light-Induced Release of the Switch Factor During Photophobic Responses of Halobacterium halobium.
Naturwissenschaften 78, 127-129 (1991). - W. Marwan, M. Alam, D: Oesterhelt:
Rotation and Switching of the Flagellar Motor Assembly in Halobacterium halobium.
J. Bacteriol 173, 1971-1977 (1991). - D. Oesterhelt, W. Marwan:
Signal Transduction in Halobacterium halobium.
In: Biology of the Chemotactic Response (J.P. Armitage, J.M. Lackie. eds.) Cambridge Univ. Press, 1990, pp. 219-239. - W. Marwan, D. Oesterhelt:
Quantitation of Photochromism of Sensory Rhodopsin-I by Computerized Tracking of Halobacterium halobium Cells.
J. Mol. Biol. 215, 277-285 (1990). - S. Sundberg, M. Alam, M. Lebert, J. Spudich, D. Oesterhelt, G. Hazelbauer:
Characterization of Mutants of Halobacterium halobium Defective in Taxis.
J. Bacteriol. 172, 2328-2335 (1990). - W. Marwan, W. Schaefer, D. Oesterhelt:
Signal Transduction in Halobacterium Depends on Fumarate.
EMBO J. 9, 355-362 (1990). - A. Blanck, D. Oesterhelt, E. Ferrando, E.S. Schegk, F. Lottspeich:
Primary Structure of Sensory Rhodopsin I, a Procaryotic Photoreceptor.
EMBO J. 8, 3963-3971 (1989) - D. Oesterhelt, W. Marwan:
Signal Transduction in Halobacteria.
NATO ASI Series, Vol H29, Receptors, Membrane Transport and Signal Transduction (A.E. Evangelopoulos et al., eds.), Springer Verlag Berlin Heidelberg 1989, pp. 282-301. - M. Alam, M. Lebert, D. Oesterhelt, G.L. Hazelbauer:
Methyl-Accepting Taxis Proteins in Halobacterium halobium.
EMBO J. 8, 631-639 (1989). - J. Otomo, W. Marwan, D. Oesterhelt:
Biosynthesis of the Two Halobacterial Light Sensors P480 and Sensory Rhodopsin and Variation in Gain of Their Signal Transduction Chains.
J. Bacteriol. 171, 2155-2159 (1989). - E.S. Schegk, D. Oesterhelt:
Isolation of a Prokaryotic Photoreceptor: Sensory Rhodopsin from Halobacteria.
EMBO J. 7, 2925-2933 (1988). - W. Marwan, M. Alam, D. Oesterhelt:
Die Geißelbewegung halophiler Bakterien.
Naturwissenschaften 74, 585-590 (1987). - [toadd] W. Marwan, P. Hegemann, D. Oesterhelt:
Single Photon Detection by an Archaebacterium.
J. Mol. Biol. 199, 663-664 (1988). - D. Oesterhelt, W. Marwan:
A Change of Membrane Potential is not a Component of the Photophobic Transduction Chain in Halobacterium halobium.
J. Bacteriol. 169, 3515-3520 (1987). - [toadd] W. Marwan, D. Oesterhelt:
Signal Formation in the Halobacterial Photophobic Response Mediated by a Fourth Retinal Protein (P475).
J. Mol. Biol. 195, 333-342 (1987). - M. Alam, D. Oesterhelt:
Purification, Reconstitution and Polymorphic Transition of Halobacterial Flagella.
J. Mol. Biol. 194, 495-499 (1987). - M. Alam, M. Claviez, D. Oesterhelt, M. Kessel:
Flagellar and Motility Behaviour of Square Bacteria.
EMBO J. 3, 2899-2903 (1984). - M. Alam, D. Oesterhelt:
Morphology, Function and Isolation of Halobacterial Flagella.
J. Mol. Biol. 176, 459-475 (1984). - W. Stoeckenius, E.K. Wolff, B. Hess
A rapid population method for action spectra applied to Halobacterium halobium.
J Bacteriol. 170, 2790-2795 (1988). - R.N. Grishanin, S.I. Bibikov, I.M. Altschuler, A.D. Kaulen, S.B. Kazimirchuk, J.P. Armitage, V.P. Skulachev
ΔΨ-mediated signalling in the bacteriorhodopsin-dependent photoresponse.
J. Bacteriol. 178, 3008-3014 (1996). - S. Hou, R.W. Larsen, D. Boudko, C.W. Riley, E. Karatan, M. Zimmer, G.W. Ordal, M. Alam
Myoglobin-like aerotaxis transducers in Archaea and Bacteria.
Nature. 403, 540-544 (2000).





