The membrane proteome of Halobacterium salinarum
Some links
- The cytosolic proteome of Halobacterium salinarum
- Membrane proteome publication:
Ch. Klein, C. Garcia-Rizo, B. Bisle, B. Scheffer, H. Zischka, F. Pfeiffer, F. Siedler, D. Oesterhelt
The membrane proteome of Halobacterium salinarum.
Proteomics 5, 180-197 (2005) - Cytosolic proteome publication:
A. Tebbe, Ch. Klein, B. Bisle, F. Siedler, B. Scheffer, C. Garcia-Rizo, J. Wolfertz, V. Hickmann, F. Pfeiffer, D. Oesterhelt
Analysis of the cytosolic proteome of Halobacterium salinarum and its implication for genome annotation.
Proteomics 5, 168-179 (2005)
Introduction
Membrane proteins play a key role in cellular processes, being involved in e.g. energy metabolism, response to environmental stimuli, and transport processes. We study membrane proteins in the halophilic archaeon Halobacterium salinarum which can grow in saturated salt solutions, can gain energy by photosynthesis using retinal as photoreceptor (more), and can sense a variety of environmental stimuli (e.g. nitrients, light, and oxygen) (more). Therefore we have analyzed the membrane proteome of this species.
There are several major groups of membrane proteins (for details see below).
* integral membrane proteins
* peripheral membrane proteins
o lipoproteins
o subunits of membrane complexes
But there are also artificial contaminants.
Result summary
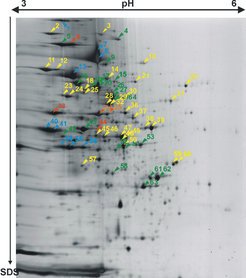
The most common strategy for proteome analysis is 2D gel electrophoresis followed by mass spectrometric protein identification (more on 2D gel electrophoresis and on mass spectrometry: MALDI-TOF, peptide mass fingerprint, and LC/MS/MS). In contrast to soluble and peripheral membrane proteins, which can be easily separated and identified by this standard proteomic techniques (Fig. 1A), the identification of integral membrane proteins by such an approach has nearly completely failed. We can attribute this failure to experimental as well as theoretical reasons:
* an irreversible precipitation of membrane proteins during isoelectric focussing that prohibits transfer to the second dimension (SDS gel electrophoresis) (Fig.4)
* the patterns generated by proteolytic cleavage are unfavourable for peptide mass fingerprint analysis. (Fig.5)
The low number of contaminants identified in the membrane preparation upon 2D gel electrophoresis is due to an optimization of the membrane preparation procedure.
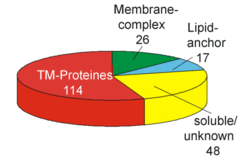
A successful approach for identfication of integral membrane proteins consists of 1D gel electrophoresis (Fig. 2A) followed by in-gel trypsin digestion and LC/MS/MS. We were able to identify 20% of the integral membrane proteins from Halobacterium salinarum by this approach. Subunits from several membrane complexes were identified, including those involved in transport processes or from the respiratory chain (Fig.3A and Fig.3B).
Result details
Fig. 1A shows a purified membrane preparation after 2D gel electrophoresis, including the identification of several peripheral membrane proteins (lipid-anchored or membrane complex subunit), but only very few integral membrane proteins (Fig. 1B). We found two reasons that preclude identification of integral membrane proteins by this technique (see Fig.4 and Fig.5).
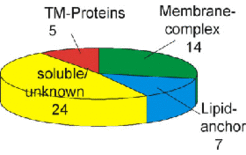
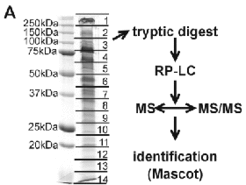
Fig. 2A shows the 1D SDS gel electrophoresis of a purified membrane preparation, which allowed the identification of more than 100 integral membrane proteins by the LC/MS/MS technique (Fig. 2B), including proteins involved in transport (Fig.4) and respiratory chain complexes (Fig.5).
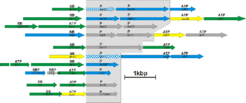
Subunits of many membrane complexes have been identified for Halobacterium salinarum. As two examples, we display operons for two types of membrane complexes:
-
ABC-type transport systems consist of three types of subunits: permeases are integral membrane proteins. The transport process is energized by cytoplasmic ATP-binding proteins. The complex may also contain a periplasmic substrate-binding protein. In Halobacterium, many of these periplasmic proteins are lipoproteins, being attached to the membrane by a lipid anchor (Fig. 3B)
-
Halobacterium salinarum can grow aerobically due to its repiratory chain. It contains homologs to all of the five complexes of the mitochondrial respiratory chain. However, there are many differences to the Halobacterium complexes to their mitochondrial counterparts (not further detailed on this page).

An experiment that proves the precipitation of membrane proteins upon isoelectric focussing by usage of fluorescently labelled proteins.

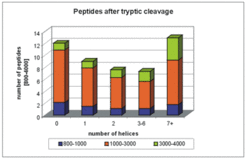
A statistical analysis of the theoretical tryptic digest shows that membrane proteins are not well suited for peptide mass fingerprint analysis. Trypsin digestion does not generate engough peptides in the mass range that is well detected upon MALDI-TOF. Large fragments (>3000 Da), which frequently are escape detection by this technique, are predominantly generated from membrane proteins.
Background information
A brief introduction to membrane proteins
Proteins may be associated with the cell membrane in several ways. Usually, when referring to membrane proteins, the focus is on integral membrane proteins . In addition, there are several groups of peripheral membrane proteins. Usually, membrane preparations also contain contaminants which can be reduced by optimization of the membrane preparation.
-
integral membrane proteins
Integral membrane proteins localized within the lipid bilayer. Most of them contain hydrophobic, membrane-spanning alpha-helices, called transmembrane domains (TMDs). -
peripheral membrane proteins
Peripheral membrane proteins (also called extrinsic membrane proteins) do not contain membrane-spanning domains but are attached to the membrane in one of two ways: -
lipoproteins
Lipoproteins are anchored to the membrane via a covalently attached lipid anchor. -
subunits of membrane complexes
Membrane complexes usually contain a core of integral membrane proteins having TMDs. Other subunits, which themselves do not have TMDs are bound to these and thus are indirectly attached to the membrane. When removed from the complex, e.g. by denaturation, they behave like soluble proteins. -
hydrophobic proteins
We noticed several hydrophobic proteins which have been excusively identified in membrane gels. These may also represent membrane proteins. They could be subunits of undescribed membrane complexes, have a lipid anchor which has not yet been detected, or have transmembrane domains which has not been found by the applied prediction programs. -
contaminants
Many proteins artificially stick to membranes although they are not functionally associated with these. Special care must be taken upon membrane preparation to reduce such contaminants.
A brief introduction to 2D gel electrophoresis
Upon 2D gel electrophoresis, proteins are separated in two dimensions. Classically, proteins are separated by charge in the first dimension. This is done in a gel stripe using "isoelectric focussing" (IEF). Proteins are separated by size in the second dimension. This is done by SDS-PAGE (Poly-Acrylamide Gel Electrophoresis in the presence of the detergent SDS). To transfer proteins from the first to the second dimension, the IEF stripe of the first dimension is first soaked in SDS buffer to re-solubilize the proteins. The stripe is then placed on top of an SDS-PAGE gel and proteins are separated in an electric field.
The transfer from the first to the second dimension fails for integral membrane proteins and largely precludes their identification by this technique.
introduction summary details background
A brief introduction to MALDI-TOF
MALDI-TOF is a method to simultaneously determine the masses of several peptides in a mixture. To permit peptide analysis in a mass spectrometer, they must be charged (ionized) and present in the gas phase, which is done by MALDI (Matrix-Assisted Laser Desorption and Ionization). Peptides are co-crystallized with a matrix, consisting of chemicals that promote transfer of charges and thus results in peptide ionization. A laser flash is used for two purposes: (a) transfer of the charged peptides into the gas phase (desorption) and (b) very precise timing, which is required to measure the "time of flight" (TOF). After desorption, peptides are accelerated by high voltage and fly towards a detector which they hit after some time. The time of flight (TOF) depends on the peptide speed, which in turn depends on their mass.
A brief introduction to PMF (peptide mass fingerprint) analysis
A peptide mass fingerprint is very specific for a given protein and permits its identification (just as a fingerprint is used for identification). Basically, a set of experimentally measured peptide fragment masses is compared to a set of theoretically computed masses. The technique depends on the specificity of proteases, which cut (digest) proteins into peptide fragments. Several proteases, e.g. trypsin, cut only after specific amino acids and thus the resulting peptide fragments and their masses can be theoretically computed. For experimental peptide mass determination, the MALDI-TOF technique is used.
A brief introduction to LC/MS/MS
In LC/MS/MS, peptides are separation by high pressure liquid chromatography (HPLC) prior to identification by mass spectrometry. HPLC reduces the complexity of the peptide mixture to permit the successive analysis step. Usage of MS/MS permits to generate a sequence tag from the fragmentation pattern of a single mass peak and thus permits protein identification from a single proteolytic peptide.







