Halobacterium salinarum
Introduction

Halobacterium salinarum is a model organism for the halophilic branch of the archaea. It is rod-shaped, motile, lives in highly saline environments (4M salt and higher), and is one of the few species known that can live in saturated salt solutions. Mass cultures of Halobacterium salinarum as shown in the pictures below can be recognized by their typical color, which originates from bacterioruberins. Halobacterium salinarum is depicted in its natural environment and as a species that colonizes salines.

It can live with light as only energy source due to the activity of the retinal protein bacteriorhodopsin, a light-driven proton pump, which has been studied in great detail and has become a paradigm of membrane proteins in general and transport proteins in particular. From this point, our focus has widened to study additional processes in which retinal proteins are involved: the energy metabolism of Halobacterium salinarum and the tactic responses with its associated signal transduction network.
Since many years, research in the department of membrane biochemistry at the Max-Planck-Institute of Biochemistry concentrates on the biology of Halobacterium salinarum.
Retinal proteins of Halobacterium salinarum
Halobacterium salinarum contains four retinal proteins , which are photosynthetic pigments with a retinal chromophore involved in light energy conversion and signal transduction. The four retinal proteins are
- bacteriorhodopsin
(Haupts et. al. (1999), Oesterhelt (1998))
- the photosynthetic pigment that permits Halobacterium to grow with light as only energy source
- a light-driven proton pump which converts light energy into a proton gradient. The energy stored in the proton gradient can be used in different ways, e.g. for generation of ATP via ATP synthase
- halorhodopsin
(Kolbe et. al. (2000), Oesterhelt (1998))
- a light-driven chloride pump that permits Halobacterium to maintain the high internal salt concentration upon growth
- sensory rhodopsin I
- involved in phototaxis, mediates the photophilic response to orange and also the photophobic response to UV light
- forms a complex with the transducer protein htrI
- sensory rhodopsin II
- involved in phototaxis, mediates the photophobic response to blue light
- forms a complex with the transducer protein htrII
Energy metabolism
Halophiles, unlike their closest relatives, the methanogenes, can grow under aerobic and anaerobic conditions. Halobacterium has three distinct systems to gain energy.
- Oxidation of various metabolites under aerobic conditions
- oxidizes pyruvate which is channeled into the tricarboxylic acid cycle using pyruvate--ferredoxin oxidoreductase ( Plaga et. al. (1992))
- Halobacterium contains all five major complexes of the respiratory chain.
- Photosynthesis
- bacteriorhodopsin, a light-driven proton pump, creates a proton gradient. ATP synthase can use the energy of the proton gradient to drive ATP synthesis.
- Fermentation of argnine
- Halobacterium generates ATP by its arginine fermentation pathway.
Response to external stimuli (signal transduction)
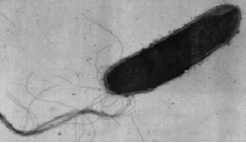
Halobacterium is a flagellated organism which shows (chemo)tactic behaviour. Besides being able to detect essential amino acids (chemotaxis) and osmotically active compounds (osmotaxis), it can respond to light (phototaxis) and can sense oxygen (aerotaxis).
The signal transduction cascade starts with the receptor/transducer, which may be composed of two distinct proteins or may be a single protein. The signal is forwarded to the switch of the flagellar motor through a two-component regulatory system consisting of the histidine kinase cheA and the response regulator cheY. During relay of the signal it is amplified and different signals are integrated (see principles of a signal transduction). Adaptation involves methylation and demethylation of the transducer proteins by cheR (methyltransferase) and cheB (a regulated methylesterase). Genome analysis shows that Halobacterium contains 18 distinct transducers, indicating that it can sense a large number of distinct stimuli. Currently, stimuli are known seven of these transducers.
- two transducers are involved in phototaxis
- photophilic response: transducer protein htrI in combination with its photoreceptor sensorhodopsin I
- photophobic response: transducer protein htrII in combination with its photoreceptor sensorhodopsin II
- two transducers are involved in aerotaxis
- aerophilic response: transducer protein htrVIII
- aerophobic response: transducer protein hemAT
( Hou et. al. (2001)) - two transducers are involved in chemotaxis towards amino acids
- chemotactic response towards Arg: soluble transducer protein car
( Storch et. al. (1999)) - chemotactic response towards Leu, Ile, Val, Met, Cys: transducer protein basT in combination with periplasmic substrate binding protein basB
( Kokoeva et. al. (2000), Kokoeva et. al. (2002)) - one transducer is involved in chemotaxis towards osmolytes
- chemotactic response towards compatible osmolytes: transducer protein cosT in combination with periplasmic substrate binding protein cosB
( Kokoeva et. al. (2002)) - transducer mpcT is a membrane potential sensor
- this is involved in BR-mediated phototaxis
( Koch et. al. (2005))
Genome analysis
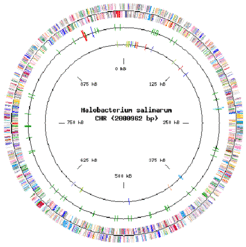
The genome of Halobacterium salinarum has been unraveled twice. The genome of Halobacterium salinarum strain R1 has been sequenced by Oesterhelt et al. (unpublished, www.halolex.mpg.de). The genome of Halobacterium salinarum strain NRC-1 has been unraveled by Ng et al.. Halobacterium has a chromosome of 2 Mb having 68% GC and a number of megaplasmids with an average of 58% GC. Strain R1 has 4 megaplasmids and strain NRC-1 is reported to have 2 megaplasmids. Approximately 2,837 proteins are encoded by the whole genome of strain R1.
References
- U. Haupts, J. Tittor, and D. Oesterhelt (1999)
-
Hou S., Freitas T., Larsen R.W., Piatibratov M., Sivozhelezov V., Yamamoto A., Meleshkevitch E.A., Zimmer M., Ordal G.W., Alam M.
Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria.
Proc. Natl. Acad. Sci. USA (2001) 98: 9353-9358 - M.V. Kokoeva, D. Oesterhelt: BasT, a Membrane-Bound Transducer Protein for Amino Acid Detection in Halobacterium salinarum. Molec. Microbiol. 35, 647-656 (2000).
- M.V. Kokoeva, K.-F. Storch, Ch. Klein, D. Oesterhelt: A Novel Mode of Sensory Transduction in Archaea: Binding Protein Mediated Chemotaxis towards Osmoprotectrants and Amino Acids in Halobacterium salinarum. EMBO J. 21, 2312-2322 (2002).
- M. Kolbe, H. Besir, L.-O. Essen, D. Oesterhelt: Structure of a Light-Driven Chloride Pump at 1.8 Å Resolution. Science, 288, 1390-1396 (2000).
-
Ng W.V., Kennedy S.P., Mahairas G.G., Berquist B., Pan M., Shukla H.D., Lasky S.R., Baliga N.S., Thorsson V., Sbrogna J., Swartzell S., Weir D., Hall J., Dahl T.A., Welti R., Goo Y.A., Leithauser B., Keller K., Cruz R., Danson M.J., Hough D.W., Maddocks D.G., Jablonski P.E., Krebs M.P., Angevine C.M., Dale H., Isenbarger T.A., Peck R.F., Pohlschroder M., Spudich J.L., Jung K.W., Alam M., Freitas T., Hou S., Daniels C.J., Dennis P.P., Omer A.D., Ebhardt H., Lowe T.M., Liang P., Riley M., Hood L., DasSarma S.:
Genome sequence of Halobacterium species NRC-1.
Proc. Natl. Acad Sci. USA (2000) 97: 12176-12181 - D. Oesterhelt (1998)
- W. Plaga, F. Lottspeich, D. Oesterhelt: Improved Purification, Crystallization and Primary Structure of Pyruvate:Ferredoxin Oxidoreductase from Halobacterium halobium. Eur. J. Biochem. 205, 391-397 (1992)
- K.F. Storch, J. Rudolph, D. Oesterhelt: Car: A Cytoplasmic Sensor Responsible for Arginine Chemotaxis in the Archaeon Halobacterium salinarum. EMBO J. 18, 1146-1158 (1999).



