Summary
Synaptic transmission is fundamental for our sensory, motor and cognitive capabilities. Like many other cellular processes, it requires a precise coordination of molecular pathways that are carried out by multiple macromolecular complexes organized in functional units. While physiological experiments, and studies of purified and reconstituted systems were essential for our understanding of the function of the individual complexes involved, they cannot resolve how the actions of individual complexes integrate. Cryo-electron tomography (cryo-ET) allows us to simultaneously image multiple protein complexes and lipids at a single nanometer resolution in their native composition, conformation and environment. Our goal is to determine the precise localization and molecular identity of complexes at mammalian synapses, and to elucidate the molecular architecture of large trans-synaptic molecular assemblies, and obtain a comprehensive picture of synaptic transmission, plasticity and synaptopathies.
Molecular architecture of trans-synaptic assemblies
Recently, we detected tripartite trans-synaptic complexes, termed subcolumns, which provide a structural link between synaptic vesicles and postsynaptic receptors. Our results showed that these complexes are characterized by a particular displacement between directly interacting components and combine in a non-uniform manner to form larger assemblies. This finding might answer the long-standing question about the mechanism of the precise alignment between neurotransmitter release sites and neuroreceptors, which is required for efficient synaptic transmission. Furthermore, we hypothesize that it provides a framework that underlies synaptic nano-columns previously described by super-resolution fluorescence imaging.

In situ, de novo structural determination of membrane-bound complexes
Applications of our template-free detection and classification procedure to in situ and isolated ER membranes containing hundreds of different protein species, allowed us to obtain de novo average structures of complexes as small as 200 kDa. This method outperformed both template-free and template-based approaches currently available in the field. Furthermore, we obtained first de novo average structures of ionotropic glutamate receptors in their physiological composition within their native environment of interacting proteins and lipids.

Structural and molecular model of synaptic vesicle tethering and priming
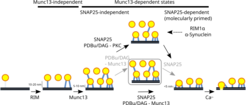
Our pioneering work on cryo-ET of mammalian synapses revealed that short pleomorphic filaments organize synaptic vesicles by tethering them to the presynaptic plasma membrane (tethers) and interlinking them to form vesicle clusters (connectors). Cryo-ET imaging of genetically and pharmacologically perturbed synapses allowed us to get data inaccessible to other methods and directly relate the synapse state to structural changes at the molecular level, which resulted in a model of synaptic vesicle tethering and priming for neurotransmitter release. Specifically, we showed that connectors are dynamic structures involved in synaptic vesicle mobilization for release and that α-Synuclein, a protein that is strongly linked to Parkinson’s disease, regulates clustering of synaptic vesicles and may modulate the neurotransmitter release. Furthermore, we found that the precise distance of a synaptic vesicle to the presynaptic plasma membrane and its tethering state are indicative of the vesicle’s progression towards fusion. While Rim1α protein is necessary for the initial tethering of synaptic vesicles at the active zone, Munc13 and SNAP-25 proteins define different tether types that localize synaptic vesicles with a single nanometer precision and are needed to bring the vesicles closer to the plasma membrane and prime them for fusion.
Computational developments
The crowded nature of cells and the large number of protein species complicates the molecular interpretation of cellular tomograms, thus making the development of novel image processing tools essential for our research. We developed software for comprehensive, template-free detection and unsupervised classification of diverse membrane bound complexes in cellular cryo-ET datasets, as well as for their morphological, topological, multi-feature statistical and spatial pattern analysis (Pyto and PySeg packages). These tools use advanced algorithms and machine learning techniques not previously used in cryo-ET (Morse theory, Affinity propagation clustering, Ripley’s functions).
In addition, we developed coordinate transformation based cryo-correlative software applicable to confocal fluorescence, transmission and scanning electron imaging, and cryo-FIB milling (3DCT package).
Publications
Papantoniou, C., Laugks U., Betzin J., Capitanio C., Ferrero J.J., Sanchez-Prieto J., Schoch S., BroseN., Baumeister W., Cooper B.H., Imig C. and Lucic V.: Munc13 and SNAP25 dependent
tethering plays a key role in synaptic vesicle priming. BioRxiv 2022.04.10.487799 (2023) in revision
Radecke, J., Seeger R., Kadkova A., Laugks U., Khosrozadeh A., Goldie K.N., Lucic V., Sørensen J.B. and Zuber B.: Morphofunctional changes at the active zone during synaptic vesicle
exocytosis, bioRxiv 2022.03.07.483217 (2023) in revision
Martinez-Sanchez, A. and Lucic V.: Detection and interpretation of cellular structures in tomograms - Segmentation, localization and spatial pattern analysis. In Foerster F. and Briegel A.,
editors, Cryo-electron Tomography, Springer, volume (2023) in revision
Lucic, V.: Computational methods for ultrastructural analysis of synaptic complexes, Current Opinion in Neurobiology 76:102611.
Zuber, B. and Lucic V.: Neurons as a model system for cryo-electron tomography, Journal of Structural Biology: X 6:100067 (2022)
Martinez-Sanchez, A., Laugks U., Kochovski Z., Papantoniou C., Zinzula L., Baumeister W. and Lucic V.: Trans-synaptic assemblies link synaptic vesicles and neuroreceptors, Science advances
7:eabe6204 (2022)
Martinez-Sanchez, A., Kochovski Z., Laugks U., Meyer Zum Alten Borgloh J., Chakraborty S., Pfeffer S., Baumeister W. and Lucic V.: Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms, Nature methods 17:209–216 (2020)
Zuber B. and Lucic V.: Molecular architecture of the presynaptic terminal, Current opinion in structural biology 54:129–138 (2019)
Ducic T., Stamenkovic S., Lai B., Andjus P. and Lucic V.: Multimodal synchrotron radiation microscopy of intact astrocytes from the hSOD1 G93A rat model of amyotrophic lateral sclerosis, Analytical chemistry 91:1460–1471 (2019)
Schrod N., Vanhecke D., Laugks U., Stein V., Fukuda Y., Schaffer M., Baumeister W. and V. Lucic: Pleomorphic linkers as ubiquitous structural organizers of vesicles in axons, PloS one
13:e0197886 (2018)
Vargas K. J., Schrod N., Davis T., Fernandez-Busnadiego R., Taguchi Y.V., Laugks U., Lucic V. and Chandra S.S.: Synucleins Have Multiple Effects on Presynaptic Architecture, Cell reports
18:161–173 (2027)
Lucic V., Fernandez-Busnadiego R., Laugks U. and W. Baumeister: Hierarchical detection and analysis of macromolecular complexes in cryo-electron tomograms using Pyto software, Journal
of structural biology 196:503–514 (2016)
Fernandez, J.-J., Laugks U., Schaffer M., Baeuerlein F.J.B., Khoshouei M., Baumeister W. and V. Lucic: Removing Contamination-Induced Reconstruction Artifacts from Cryo-electron Tomograms, Biophys J 110:850–859 (2016)
Arnold J., Mahamid J., Lucic V., de Marco A., Fernandez J.J., Laugks T., Mayer T., Hyman A.A., Baumeister W. and Plitzko J.M.: Site-Specific Cryo-focused Ion Beam Sample Preparation Guided by 3D Correlative Microscopy, Biophys J 110:860–869 (2016)
Fukuda Y., Laugks U., Lucic V., Baumeister W. and Danev R.: Electron cryotomography of vitrified cells with a Volta phase plate, J Struct Biol 190:143–154 (2015)
Lucic V.: Neuroscience: towards unified vesicle endocytosis, Nature 515:207–208 (2014)
Fukuda Y., Schrod N., Schaffer M., Feng L.R., Baumeister W. and Lucic V.: Coordinate transformation based cryo-correlative methods for electron tomography and focused ion beam milling, Ultramicroscopy 143:15–23 (2014)
Martinez-Sanchez A., Garcia I., Asano S., Lucic V. and Fernandez J.J.: Robust membrane detection based on tensor voting for electron tomography, J Struct Biol 186:49–61 (2014)
Lucic V., Rigort A. and Baumeister W.: Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202:407–419 (2013)
Lucic V. and Baumeister W.: 3D Electron Microscopy Based on Cryo-Electron Tomography, In G. C. K. Roberts, editor, Encyclopedia of Biophysics, Springer-Verlag Berlin Heidelberg, 7–10 (2013)
Fernandez-Busnadiego R. and Lucic V.: The Cell at Molecular Resolution: Principles and Applications af Cryo-Electron Tomography, In F. G. Wouterlood, editor, Cellular Imaging Techniques for Neuroscience and Beyond, Elsevier, 141–183 (2012)
Lucic V., Forster F. and W. Baumeister W.: Studying the macromolecular machinery of cells in situ by cryo-electron tomography. In E. Egelman, and P. Schwille, editors, Comprehensive Biophysics 2, Biophysical Techniques for Characterization of Cells, Elsevier B. V. Academic Press, Oxford, 59–89 (2012)
Fernandez-Busnadiego R., Schrod N., Kochovski Z., Asano S., Vanhecke D., Baumeister W. and V. Lucic: Insights into the molecular organization of the neuron by cryo-electron tomography, J Electron Microsc (Tokyo) 60 Suppl 1:S137–S148 (2011)
Vanhecke D., Asano S., Kochovski Z., Fernandez-Busnadiego R., Schrod N., Baumeister W. and V. Lucic: Cryo-electron tomography: methodology, developments and biological applications, J Microsc 242:221–227 (2011)
Fernandez-Busnadiego R., Zuber B., Maurer U.E., Cyrklaff M., Baumeister W. and Lucic V.: Quantitative analysis of the native presynaptic cytomatrix by cryoelectron tomography, J Cell Biol 188:145–56 (2010)
Lucic V., Leis A. and Baumeister W.: Cryo-electron tomography of cells: connecting structure and function, Histochem Cell Biol 130:185–96 (2008)
Beck M., Lucic V., Foerster F., Baumeister W. and Medalia O.: Snapshots of nuclear pore complexes in action captured by cryo-electron tomography, Nature 449:611–5 (2007)
Lucic V., Kossel A.H., Yang T., Bonhoeffer T., Baumeister W. and Sartori A.: Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography, J Struct Biol 160:146–56 (2007)
Fernandez J.-J., Li S. and Lucic V.: Three-dimensional anisotropic noise reduction with automated parameter tuning: Application to electron cryotomography, In Borrajo D.; Castillo L.; Corchado J.M., editor, Current topics in artificial inteligence, Springer Verlag, volume 4788 of Lecture notes in computer science series. Lecture notes in artificial intelligence subseries., 60–69 (2007)
Fernandez J.-J., Li S. and Lucic V.: Structure-preserving noise reduction in biological imaging, In Corchado E.; Corchado J.M.; Abraham A, editor, Innovation in hybrid intelligent systems, Springer Verlag, volume 44 of Advances in soft computing, 385–392 (2007)
Korkin D., Davis F.P., Alber F., Luong T., Shen M.Y., Lucic V., Kennedy M.B. and Sali A.: Structural modeling of protein interactions by analogy: application to PSD-95, PLoS Comput Biol 2:e153 (2006)
Lucic V., Foerster F. and Baumeister W.: Structural studies by electron tomography: from cells to molecules, Annu Rev Biochem 74:833–65 (2005)
Lucic V., Yang T., Schweikert G., Forster F. and Baumeister W.: Morphological characterization of molecular complexes present in the synaptic cleft, Structure 13:423–34 (2005)
Lucic V. and Baumeister W.: Neuroscience. Monte Carlo places strong odds on ectopic release, Science 309:387–8 (2005)


