The Macromolecular Shredder for RNA in the Cell Nucleus
Much in the same way as we use shredders to destroy documents that are no longer useful or that contain potentially damaging information, cells use molecular machines to degrade unwanted or defective macromolecules. Scientists of the Max Planck Institute of Biochemistry (MPIB) in Martinsried near Munich, Germany, have now shown how the nuclear compartment of the cell uses a specific version of the RNA exosome, a macromolecular machine responsible for the degradation as well as the biogenesis of ribonucleic acids (RNAs). RNAs are ubiquitous and abundant molecules with multiple functions in the cell. One of their functions is, for example, to permit translation of the genomic information into proteins. The study has now been published in the journal Nature.
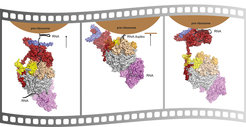
Any errors that occur during the synthesis of RNA molecules or unwanted accumulation of RNAs can be damaging to the cell. The elimination of defective RNAs or of RNAs that are no longer needed are therefore key steps in the metabolism of a cell. The exosome, a multi-protein complex, is a key machine that shreds RNA into pieces. In addition, the exosome also processes certain RNA molecules into their mature form. In a study two years ago, scientists in the Research Department ‘Structural Cell Biology’ headed by Elena Conti unveiled the X-ray structure of the exosome core complex. The multi-protein complex consists of nine proteins that form a central substrate channel that ends in the protein Rrp44, the exosome RNA degrading center.
Specific shredders for each compartment of the cell
Different cellular compartments, such as the nucleus or the cytoplasm, have their own specific versions of a larger exosome complex bound to specific helper proteins. The MPIB scientists could now reveal how the exosome in the nucleus works together with two protein-subunits called Rrp6 and Rrp47, which are specific only for RNA substrates of the nucleus. “We could show that the cell has multiple possible paths of degrading nuclear RNA,” explains Debora Makino, one of the authors of the study. One of the pathways leads the RNA substrate into direct degradation by Rrp6 and/or Rrp44, and the other guides the RNA into the processive degradation by Rrp44 via the core RNA exosome channel path. “In this manner, the cell can degrade RNA substrates either completely or trim them precisely when needed,” says Benjamin Schuch, the other author of the study.
Future research will uncover further RNA processing mechanisms involving the core exosome and its various auxiliary proteins, protein complexes, and RNA substrates located throughout all cell compartments.
Original publication:
D.L. Makino*, B. Schuch*, E. Stegmann, M. Baumgärtner, C. Basquin and E. Conti: RNA degradation paths in a 12-subunit nuclear exosome complex. Nature, July 29, 2015
DOI: 10.1038/nature14865












