Anatomy of a nematode - molecule by molecule
A new dimension in cryo-electron tomography: Researchers, led by Jürgen Plitzko at the MPI of Biochemistry, improve workflows to enable the anatomical imaging of multicellular organisms at molecular resolution.
Researchers led by Jürgen Plitzko at the Max Planck Institute (MPI) of Biochemistry have succeeded in significantly simplifying a key step in the research routine of scientists in cryo-electron tomography (cryo-ET). Thanks to their newly developed method, it is now possible for the first time to obtain a series of sections from the model organism C. elegans and image them using cryo-ET. This opens the door to the anatomical study of multicellular organisms and tissue samples at molecular resolution. The results are published in the journal Nature Methods.
The study of human anatomy in healthy and diseased states is a crucial part of modern medicine. Studying the anatomy of individual cells at the molecular level, however, requires complex imaging methods with a very high resolution.
Cryo-electron tomography (cryo-ET) is a method that allows for the internal structure of cells to be visualized down to individual proteins. Similar to computed tomography (CT), which can be used to examine the human body, cryo-ET also involves transilluminating a sample from different directions. The resulting image information can then be used to obtain a three-dimensional reconstruction of the examined object. In contrast to CT, in which people can simply lie down in a machine, cryo-ET requires a complex sample preparation that crucially impacts the quality of the data obtained.
The samples used must be extremely thin. Even a single human cell is still too thick and must first be thinned into a slice called a lamella for this method. This slice is then only about one four-hundredth of the thickness of a human hair. However, before the sample can be thinned, it must first be preserved by 'shock freezing' it, so to speak. In this special type of freezing, called vitrification, the sample is frozen so quickly that no ice crystals form. These would otherwise destroy the delicate structures inside the sample.
"Through technological advances in recent years, it is already possible to visualize small structures in individual human cells. However, such investigations have hardly been possible for small multicellular organisms or entire tissues until now," explains Christoph Kaiser, co-first author of the study.
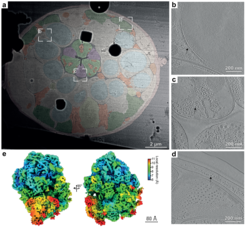
The researchers have now succeeded in optimizing the previously extremely complex sample preparation process so that it can be carried out more efficiently and with a higher success rate. In addition, it now provides not just one, but several lamellae from one sample. To do this, the scientists vitrified freshly hatched larvae of the nematode C. elegans and cut out an entire larva in a cuboid block of ice. This block was then repeatedly attached to a sample carrier and a slice was cut off. In total, the researchers succeeded in obtaining 32 slices from that individual. This procedure is illustrated in the figure.

"I think the most remarkable thing about our new method is that we have to discard much less biological material than before. Previously, we could only examine one, or very few, sections per organism or cell and the rest had to be discarded. As a result, we now obtain more information in its anatomical context, while still preserving the molecular details," says Oda Schiøtz, co-first author of the study.
To illustrate the enormous potential of their newly developed method, the researchers show high-resolution images of various anatomical structures, such as muscles, nerves and intestine, in their near-native state and physiological context. In addition, they were able to reconstruct the structure of two important cellular protein complexes from these data: the ribosome, the cellular machine for protein production, and microtubules, which are an important component of the cellular skeleton and stabilize our cells.
"Our new method provides groundbreaking insight into the molecular structure of large cells, small organisms, or whole tissue samples. Our next goal will be to make this method even faster and apply it to a variety of biological samples," says Sven Klumpe, co-corresponding author of the study. Jürgen Plitzko, head of the CryoEM Technology research group and co-corresponding author of the study, adds: "On the one hand, the method can help to answer fundamental biological questions in cell and tissue biology, but on the other hand it also provides us with more detailed insights into the molecular processes involved in diseases, such as those caused by viral or bacterial infections."
[tb]
Dictionary of Cryo-EM Technology research group:
Cryo-electron tomography (Cryo-ET): In cryo-electron tomography, two-dimensional images are recorded from different tilt angles using a special microscope (transmission electron microscope). This must be carried out at a temperature below -160°C and requires extremely thin samples. The sample’s 3D structure can then be reconstructed from the images.
Vitrification: Vitrification describes a process in which water is frozen extremely fast in just a few milliseconds. Ideally, the formation of ice crystals can be completely prevented.
Lamella/Lamellae: As extremely thin samples are required for cryo-electron tomography, the tissues or cells to be examined must first be thinned out. What remains are the so-called lamellae, which are used for cryo-ET.
Ribosome: Ribosomes are complexes consisting of proteins and RNA. In our cells, they are responsible for producing proteins.
Microtubules: Microtubules are tubular protein complexes that are responsible for the stability of the cell. Together with other proteins, they are also involved in transport processes such as the distribution of genetic information during cell division.


